Mycobacterium tuberculosis Hip1 modulates macrophage responses through proteolysis of GroEL2
- PMID: 24830429
- PMCID: PMC4022732
- DOI: 10.1371/journal.ppat.1004132
Mycobacterium tuberculosis Hip1 modulates macrophage responses through proteolysis of GroEL2
Abstract
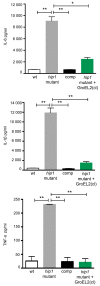
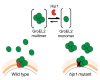
Mycobacterium tuberculosis (Mtb) employs multiple strategies to evade host immune responses and persist within macrophages. We have previously shown that the cell envelope-associated Mtb serine hydrolase, Hip1, prevents robust macrophage activation and dampens host pro-inflammatory responses, allowing Mtb to delay immune detection and accelerate disease progression. We now provide key mechanistic insights into the molecular and biochemical basis of Hip1 function. We establish that Hip1 is a serine protease with activity against protein and peptide substrates. Further, we show that the Mtb GroEL2 protein is a direct substrate of Hip1 protease activity. Cleavage of GroEL2 is specifically inhibited by serine protease inhibitors. We mapped the cleavage site within the N-terminus of GroEL2 and confirmed that this site is required for proteolysis of GroEL2 during Mtb growth. Interestingly, we discovered that Hip1-mediated cleavage of GroEL2 converts the protein from a multimeric to a monomeric form. Moreover, ectopic expression of cleaved GroEL2 monomers into the hip1 mutant complemented the hyperinflammatory phenotype of the hip1 mutant and restored wild type levels of cytokine responses in infected macrophages. Our studies point to Hip1-dependent proteolysis as a novel regulatory mechanism that helps Mtb respond rapidly to changing host immune environments during infection. These findings position Hip1 as an attractive target for inhibition for developing immunomodulatory therapeutics against Mtb.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Mycobacterium tuberculosis GroEL2 Modulates Dendritic Cell Responses.Infect Immun. 2018 Jan 22;86(2):e00387-17. doi: 10.1128/IAI.00387-17. Print 2018 Feb. Infect Immun. 2018. PMID: 29133346 Free PMC article.
-
Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).Biochemistry. 2017 May 2;56(17):2304-2314. doi: 10.1021/acs.biochem.6b01066. Epub 2017 Apr 7. Biochemistry. 2017. PMID: 28346784 Free PMC article.
-
Mycobacterium tuberculosis Hip1 dampens macrophage proinflammatory responses by limiting toll-like receptor 2 activation.Infect Immun. 2011 Dec;79(12):4828-38. doi: 10.1128/IAI.05574-11. Epub 2011 Sep 26. Infect Immun. 2011. PMID: 21947769 Free PMC article.
-
The Role of mRNA Alternative Splicing in Macrophages Infected with Mycobacterium tuberculosis: A Field Needing to Be Discovered.Molecules. 2024 Apr 16;29(8):1798. doi: 10.3390/molecules29081798. Molecules. 2024. PMID: 38675618 Free PMC article. Review.
-
Trans-species communication in the Mycobacterium tuberculosis-infected macrophage.Immunol Rev. 2015 Mar;264(1):233-48. doi: 10.1111/imr.12254. Immunol Rev. 2015. PMID: 25703563 Free PMC article. Review.
Cited by
-
MTS1338, A Small Mycobacterium tuberculosis RNA, Regulates Transcriptional Shifts Consistent With Bacterial Adaptation for Entering Into Dormancy and Survival Within Host Macrophages.Front Cell Infect Microbiol. 2019 Nov 26;9:405. doi: 10.3389/fcimb.2019.00405. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31850238 Free PMC article.
-
Systematic Survey of Serine Hydrolase Activity in Mycobacterium tuberculosis Defines Changes Associated with Persistence.Cell Chem Biol. 2016 Feb 18;23(2):290-298. doi: 10.1016/j.chembiol.2016.01.003. Epub 2016 Feb 4. Cell Chem Biol. 2016. PMID: 26853625 Free PMC article.
-
Design of Selective Substrates and Activity-Based Probes for Hydrolase Important for Pathogenesis 1 (HIP1) from Mycobacterium tuberculosis.ACS Infect Dis. 2016 Nov 11;2(11):807-815. doi: 10.1021/acsinfecdis.6b00092. Epub 2016 Jul 15. ACS Infect Dis. 2016. PMID: 27739665 Free PMC article.
-
Mycobacterium tuberculosis inhibits the NLRP3 inflammasome activation via its phosphokinase PknF.PLoS Pathog. 2021 Jul 29;17(7):e1009712. doi: 10.1371/journal.ppat.1009712. eCollection 2021 Jul. PLoS Pathog. 2021. PMID: 34324582 Free PMC article.
-
Mycobacterium tuberculosis GroEL2 Modulates Dendritic Cell Responses.Infect Immun. 2018 Jan 22;86(2):e00387-17. doi: 10.1128/IAI.00387-17. Print 2018 Feb. Infect Immun. 2018. PMID: 29133346 Free PMC article.
References
-
- Philips JA, Ernst JD (2012) Tuberculosis pathogenesis and immunity. Annu Rev Pathol 7: 353–384. - PubMed
-
- Russell DG (2001) Mycobacterium tuberculosis: here today, here tomorrow. Nature Reviews 2: 569–577. - PubMed
-
- Almeida Da Silva PE, Palomino JC (2011) Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 66: 1417–1430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

