The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components
- PMID: 24830558
- PMCID: PMC4057749
- DOI: 10.3390/ijms15058570
The G protein-coupled receptor heterodimer network (GPCR-HetNet) and its hub components
Abstract
G protein-coupled receptors (GPCRs) oligomerization has emerged as a vital characteristic of receptor structure. Substantial experimental evidence supports the existence of GPCR-GPCR interactions in a coordinated and cooperative manner. However, despite the current development of experimental techniques for large-scale detection of GPCR heteromers, in order to understand their connectivity it is necessary to develop novel tools to study the global heteroreceptor networks. To provide insight into the overall topology of the GPCR heteromers and identify key players, a collective interaction network was constructed. Experimental interaction data for each of the individual human GPCR protomers was obtained manually from the STRING and SCOPUS databases. The interaction data were used to build and analyze the network using Cytoscape software. The network was treated as undirected throughout the study. It is comprised of 156 nodes, 260 edges and has a scale-free topology. Connectivity analysis reveals a significant dominance of intrafamily versus interfamily connections. Most of the receptors within the network are linked to each other by a small number of edges. DRD2, OPRM, ADRB2, AA2AR, AA1R, OPRK, OPRD and GHSR are identified as hubs. In a network representation 10 modules/clusters also appear as a highly interconnected group of nodes. Information on this GPCR network can improve our understanding of molecular integration. GPCR-HetNet has been implemented in Java and is freely available at http://www.iiia.csic.es/~ismel/GPCR-Nets/index.html.
Figures
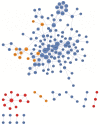



Similar articles
-
Requirements and ontology for a G protein-coupled receptor oligomerization knowledge base.BMC Bioinformatics. 2007 May 30;8:177. doi: 10.1186/1471-2105-8-177. BMC Bioinformatics. 2007. PMID: 17537266 Free PMC article.
-
Structure modeling of all identified G protein-coupled receptors in the human genome.PLoS Comput Biol. 2006 Feb;2(2):e13. doi: 10.1371/journal.pcbi.0020013. Epub 2006 Feb 17. PLoS Comput Biol. 2006. PMID: 16485037 Free PMC article.
-
Extended Human G-Protein Coupled Receptor Network: Cell-Type-Specific Analysis of G-Protein Coupled Receptor Signaling Pathways.J Proteome Res. 2020 Jan 3;19(1):511-524. doi: 10.1021/acs.jproteome.9b00754. Epub 2019 Dec 12. J Proteome Res. 2020. PMID: 31774292
-
GPCR & company: databases and servers for GPCRs and interacting partners.Adv Exp Med Biol. 2014;796:185-204. doi: 10.1007/978-94-007-7423-0_9. Adv Exp Med Biol. 2014. PMID: 24158806 Review.
-
Prediction and targeting of GPCR oligomer interfaces.Prog Mol Biol Transl Sci. 2020;169:105-149. doi: 10.1016/bs.pmbts.2019.11.007. Epub 2020 Jan 6. Prog Mol Biol Transl Sci. 2020. PMID: 31952684 Review.
Cited by
-
Multiple D2 heteroreceptor complexes: new targets for treatment of schizophrenia.Ther Adv Psychopharmacol. 2016 Apr;6(2):77-94. doi: 10.1177/2045125316637570. Epub 2016 Mar 10. Ther Adv Psychopharmacol. 2016. PMID: 27141290 Free PMC article. Review.
-
Dopamine D3 Receptor Antagonists as Potential Therapeutics for the Treatment of Neurological Diseases.Front Neurosci. 2016 Oct 5;10:451. doi: 10.3389/fnins.2016.00451. eCollection 2016. Front Neurosci. 2016. PMID: 27761108 Free PMC article. Review.
-
Oligomeric Receptor Complexes and Their Allosteric Receptor-Receptor Interactions in the Plasma Membrane Represent a New Biological Principle for Integration of Signals in the CNS.Front Mol Neurosci. 2019 Sep 25;12:230. doi: 10.3389/fnmol.2019.00230. eCollection 2019. Front Mol Neurosci. 2019. PMID: 31607863 Free PMC article.
-
The World of GPCR dimers - Mapping dopamine receptor D2 homodimers in different activation states and configuration arrangements.Comput Struct Biotechnol J. 2023 Sep 3;21:4336-4353. doi: 10.1016/j.csbj.2023.08.032. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37711187 Free PMC article.
-
Neuroprotection afforded by targeting G protein-coupled receptors in heteromers and by heteromer-selective drugs.Front Pharmacol. 2023 Jul 13;14:1222158. doi: 10.3389/fphar.2023.1222158. eCollection 2023. Front Pharmacol. 2023. PMID: 37521478 Free PMC article. Review.
References
-
- Xia Y., Yu H., Jansen R., Seringhaus M., Baxter S., Greenbaum D., Zhao H., Gerstein M. Analyzing cellular biochemistry in terms of molecular networks. Ann. Rev. Biochem. 2004;73:1051–1087. - PubMed
-
- Borroto-Escuela D.O., Agnati L.F., Fuxe K., Ciruela F. Muscarinic acetylcholine receptor-interacting proteins (mAChRIPs): Targeting the receptorsome. Curr. Drug Targets. 2012;13:53–71. - PubMed
-
- Borroto-Escuela D.O., Correia P.A., Romero-Fernandez W., Narvaez M., Fuxe K., Ciruela F., Garriga P. Muscarinic receptor family interacting proteins: Role in receptor function. J. Neurosci. Methods. 2011;195:161–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

