The role of mRNA splicing in prostate cancer
- PMID: 24830689
- PMCID: PMC4104073
- DOI: 10.4103/1008-682X.127825
The role of mRNA splicing in prostate cancer
Abstract
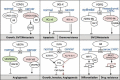
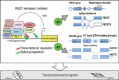
Alternative splicing (AS) is a crucial step in gene expression. It is subject to intricate regulation, and its deregulation in cancer can lead to a wide array of neoplastic phenotypes. A large body of evidence implicates splice isoforms in most if not all hallmarks of cancer, including growth, apoptosis, invasion and metastasis, angiogenesis, and metabolism. AS has important clinical implications since it can be manipulated therapeutically to treat cancer and represents a mechanism of resistance to therapy. In prostate cancer (PCa) AS also plays a prominent role and this review will summarize the current knowledge of alternatively spliced genes with important functional consequences. We will highlight accumulating evidence on AS of the components of the two critical pathways in PCa: androgen receptor (AR) and phosphoinositide 3-kinase (PI3K). These observations together with data on dysregulation of splice factors in PCa suggest that AR and PI3K pathways may be interconnected with previously unappreciated splicing regulatory networks. In addition, we will discuss several lines of evidence implicating splicing regulation in the development of the castration resistance.
Figures

References
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

