The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay
- PMID: 24830819
- PMCID: PMC4022661
- DOI: 10.1371/journal.pone.0097408
The validation and clinical implementation of BRCAplus: a comprehensive high-risk breast cancer diagnostic assay
Erratum in
- PLoS One. 2014;9(9):e110156
Abstract
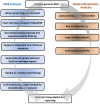
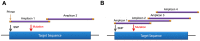
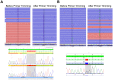
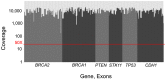
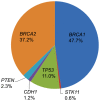
Breast cancer is the most commonly diagnosed cancer in women, with 10% of disease attributed to hereditary factors. Although BRCA1 and BRCA2 account for a high percentage of hereditary cases, there are more than 25 susceptibility genes that differentially impact the risk for breast cancer. Traditionally, germline testing for breast cancer was performed by Sanger dideoxy terminator sequencing in a reflexive manner, beginning with BRCA1 and BRCA2. The introduction of next-generation sequencing (NGS) has enabled the simultaneous testing of all genes implicated in breast cancer resulting in diagnostic labs offering large, comprehensive gene panels. However, some physicians prefer to only test for those genes in which established surveillance and treatment protocol exists. The NGS based BRCAplus test utilizes a custom tiled PCR based target enrichment design and bioinformatics pipeline coupled with array comparative genomic hybridization (aCGH) to identify mutations in the six high-risk genes: BRCA1, BRCA2, PTEN, TP53, CDH1, and STK11. Validation of the assay with 250 previously characterized samples resulted in 100% detection of 3,025 known variants and analytical specificity of 99.99%. Analysis of the clinical performance of the first 3,000 BRCAplus samples referred for testing revealed an average coverage greater than 9,000X per target base pair resulting in excellent specificity and the sensitivity to detect low level mosaicism and allele-drop out. The unique design of the assay enabled the detection of pathogenic mutations missed by previous testing. With the abundance of NGS diagnostic tests being released, it is essential that clinicians understand the advantages and limitations of different test designs.
Conflict of interest statement
Figures


References
-
- Campeau PM, Foulkes WD, Tischkowitz MD (2008) Hereditary breast cancer: New genetic developments, new therapeutic avenues. Hum Genet 124(1): 31–42. - PubMed
-
- Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, et al... (eds.) (2013) SEER Cancer Statistics Review, 1975–2010. Bethesda, MD: National Cancer Institute.
-
- King MC, Marks JH, Mandell JB (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302(5645): 643–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

