Global genetic response in a cancer cell: self-organized coherent expression dynamics
- PMID: 24831017
- PMCID: PMC4022610
- DOI: 10.1371/journal.pone.0097411
Global genetic response in a cancer cell: self-organized coherent expression dynamics
Erratum in
- PLoS One. 2014;9(8):e105491
Abstract
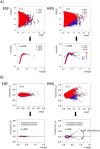
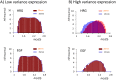
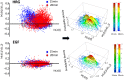
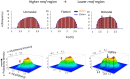
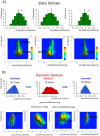
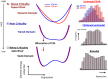
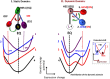
Understanding the basic mechanism of the spatio-temporal self-control of genome-wide gene expression engaged with the complex epigenetic molecular assembly is one of major challenges in current biological science. In this study, the genome-wide dynamical profile of gene expression was analyzed for MCF-7 breast cancer cells induced by two distinct ErbB receptor ligands: epidermal growth factor (EGF) and heregulin (HRG), which drive cell proliferation and differentiation, respectively. We focused our attention to elucidate how global genetic responses emerge and to decipher what is an underlying principle for dynamic self-control of genome-wide gene expression. The whole mRNA expression was classified into about a hundred groups according to the root mean square fluctuation (rmsf). These expression groups showed characteristic time-dependent correlations, indicating the existence of collective behaviors on the ensemble of genes with respect to mRNA expression and also to temporal changes in expression. All-or-none responses were observed for HRG and EGF (biphasic statistics) at around 10-20 min. The emergence of time-dependent collective behaviors of expression occurred through bifurcation of a coherent expression state (CES). In the ensemble of mRNA expression, the self-organized CESs reveals distinct characteristic expression domains for biphasic statistics, which exhibits notably the presence of criticality in the expression profile as a route for genomic transition. In time-dependent changes in the expression domains, the dynamics of CES reveals that the temporal development of the characteristic domains is characterized as autonomous bistable switch, which exhibits dynamic criticality (the temporal development of criticality) in the genome-wide coherent expression dynamics. It is expected that elucidation of the biophysical origin for such critical behavior sheds light on the underlying mechanism of the control of whole genome.
Conflict of interest statement
Figures


References
-
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 131: 861–872. - PubMed
-
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. - PubMed
-
- Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TTJP, et al. (2005) Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S-cerevisiae stationary phase exit. Mol Cell 18: 171–183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

