How do you solve a problem like Letharia? A new look at cryptic species in lichen-forming fungi using Bayesian clustering and SNPs from multilocus sequence data
- PMID: 24831224
- PMCID: PMC4022584
- DOI: 10.1371/journal.pone.0097556
How do you solve a problem like Letharia? A new look at cryptic species in lichen-forming fungi using Bayesian clustering and SNPs from multilocus sequence data
Abstract
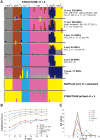
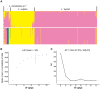
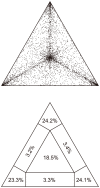
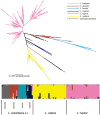
The inclusion of molecular data is increasingly an integral part of studies assessing species boundaries. Analyses based on predefined groups may obscure patterns of differentiation, and population assignment tests provide an alternative for identifying population structure and barriers to gene flow. In this study, we apply population assignment tests implemented in the programs STRUCTURE and BAPS to single nucleotide polymorphisms from DNA sequence data generated for three previous studies of the lichenized fungal genus Letharia. Previous molecular work employing a gene genealogical approach circumscribed six species-level lineages within the genus, four putative lineages within the nominal taxon L. columbiana (Nutt.) J.W. Thomson and two sorediate lineages. We show that Bayesian clustering implemented in the program STRUCTURE was generally able to recover the same six putative Letharia lineages. Population assignments were largely consistent across a range of scenarios, including: extensive amounts of missing data, the exclusion of SNPs from variable markers, and inferences based on SNPs from as few as three gene regions. While our study provided additional evidence corroborating the six candidate Letharia species, the equivalence of these genetic clusters with species-level lineages is uncertain due, in part, to limited phylogenetic signal. Furthermore, both the BAPS analysis and the ad hoc ΔK statistic from results of the STRUCTURE analysis suggest that population structure can possibly be captured with fewer genetic groups. Our findings also suggest that uneven sampling across taxa may be responsible for the contrasting inferences of population substructure. Our results consistently supported two distinct sorediate groups, 'L. lupina' and L. vulpina, and subtle morphological differences support this distinction. Similarly, the putative apotheciate species 'L. lucida' was also consistently supported as a distinct genetic cluster. However, additional studies will be required to elucidate the relationships of other L. columbiana s.l. populations with the two sorediate genetic clusters.
Conflict of interest statement
Figures


References
-
- Mayr E (1970) Populations, species, and evolution. Cambridge, Massachusetts: Belknap Press of Harvard University Press.
-
- Carstens BC, Pelletier TA, Reid NM, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22: 4369–4383. - PubMed
-
- Yeates DK, Seago A, Nelson L, Cameron SL, Joseph LEO, et al. (2011) Integrative taxonomy, or iterative taxonomy? Syst Entomol 36: 209–217.
-
- Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C (2012) Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol 9: 480–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

