Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated
- PMID: 24831516
- PMCID: PMC4022528
- DOI: 10.1371/journal.pntd.0002856
Hemoglobin uptake by Paracoccidioides spp. is receptor-mediated
Abstract
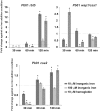
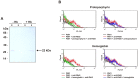
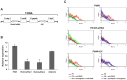
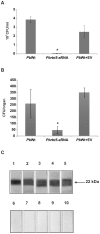
Iron is essential for the proliferation of fungal pathogens during infection. The availability of iron is limited due to its association with host proteins. Fungal pathogens have evolved different mechanisms to acquire iron from host; however, little is known regarding how Paracoccidioides species incorporate and metabolize this ion. In this work, host iron sources that are used by Paracoccidioides spp. were investigated. Robust fungal growth in the presence of the iron-containing molecules hemin and hemoglobin was observed. Paracoccidioides spp. present hemolytic activity and have the ability to internalize a protoporphyrin ring. Using real-time PCR and nanoUPLC-MSE proteomic approaches, fungal growth in the presence of hemoglobin was shown to result in the positive regulation of transcripts that encode putative hemoglobin receptors, in addition to the induction of proteins that are required for amino acid metabolism and vacuolar protein degradation. In fact, one hemoglobin receptor ortholog, Rbt5, was identified as a surface GPI-anchored protein that recognized hemin, protoporphyrin and hemoglobin in vitro. Antisense RNA technology and Agrobacterium tumefaciens-mediated transformation were used to generate mitotically stable Pbrbt5 mutants. The knockdown strain had a lower survival inside macrophages and in mouse spleen when compared with the parental strain, which suggested that Rbt5 could act as a virulence factor. In summary, our data indicate that Paracoccidioides spp. can use hemoglobin as an iron source most likely through receptor-mediated pathways that might be relevant for pathogenic mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Kaplan CD, Kaplan J (2009) Iron acquisition and transcriptional regulation. Chem Rev 109: 4536–4552. - PubMed
-
- Nevitt T (2011) War-Fe-re: iron at the core of fungal virulence and host immunity. Biometals 24: 547–558. - PubMed
-
- Ramakrishna G, Rooke TW, Cooper LT (2003) Iron and peripheral arterial disease: revisiting the iron hypothesis in a different light. Vasc Med 8: 203–210. - PubMed
-
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C (2010) Two to tango: regulation of Mammalian iron metabolism. Cell 142: 24–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

