A new restriction endonuclease-based method for highly-specific detection of DNA targets from methicillin-resistant Staphylococcus aureus
- PMID: 24831802
- PMCID: PMC4022673
- DOI: 10.1371/journal.pone.0097826
A new restriction endonuclease-based method for highly-specific detection of DNA targets from methicillin-resistant Staphylococcus aureus
Abstract
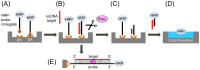
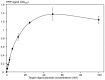
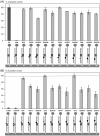
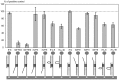
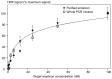
PCR multiplexing has proven to be challenging, and thus has provided limited means for pathogen genotyping. We developed a new approach for analysis of PCR amplicons based on restriction endonuclease digestion. The first stage of the restriction enzyme assay is hybridization of a target DNA to immobilized complementary oligonucleotide probes that carry a molecular marker, horseradish peroxidase (HRP). At the second stage, a target-specific restriction enzyme is added, cleaving the target-probe duplex at the corresponding restriction site and releasing the HRP marker into solution, where it is quantified colorimetrically. The assay was tested for detection of the methicillin-resistant Staphylococcus aureus (MRSA) pathogen, using the mecA gene as a target. Calibration curves indicated that the limit of detection for both target oligonucleotide and PCR amplicon was approximately 1 nM. Sequences of target oligonucleotides were altered to demonstrate that (i) any mutation of the restriction site reduced the signal to zero; (ii) double and triple point mutations of sequences flanking the restriction site reduced restriction to 50-80% of the positive control; and (iii) a minimum of a 16-bp target-probe dsDNA hybrid was required for significant cleavage. Further experiments showed that the assay could detect the mecA amplicon from an unpurified PCR mixture with detection limits similar to those with standard fluorescence-based qPCR. Furthermore, addition of a large excess of heterologous genomic DNA did not affect amplicon detection. Specificity of the assay is very high because it involves two biorecognition steps. The proposed assay is low-cost and can be completed in less than 1 hour. Thus, we have demonstrated an efficient new approach for pathogen detection and amplicon genotyping in conjunction with various end-point and qPCR applications. The restriction enzyme assay may also be used for parallel analysis of multiple different amplicons from the same unpurified mixture in broad-range PCR applications.
Conflict of interest statement
Figures


References
-
- Wolk D, Mitchell S, Patel R (2001) Principles of molecular microbiology testing methods. Infectious Disease Clinics of North America 15: 1157–1204. - PubMed
-
- Barken KB, Haagensen JAJ, Tolker-Nielsen T (2007) Advances in nucleic acid-based diagnostics of bacterial infections. Clinica Chimica Acta 384: 1–11. - PubMed
-
- Ghindilis AL, Smith MW, Schwarzkopf KR, Roth KM, Peyvan K, et al. (2007) CombiMatrix oligonucleotide arrays: genotyping and gene expression assays employing electrochemical detection. Biosens Bioelectron 22: 1853–1860. - PubMed
-
- Lazcka O, Campo F, Munoz FX (2007) Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron 22: 1205–1217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

