DNA methylation characteristics of primary melanomas with distinct biological behaviour
- PMID: 24832207
- PMCID: PMC4022506
- DOI: 10.1371/journal.pone.0096612
DNA methylation characteristics of primary melanomas with distinct biological behaviour
Abstract
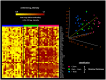
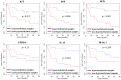
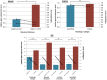
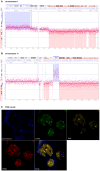
In melanoma, the presence of promoter related hypermethylation has previously been reported, however, no methylation-based distinction has been drawn among the diverse melanoma subtypes. Here, we investigated DNA methylation changes associated with melanoma progression and links between methylation patterns and other types of somatic alterations, including the most frequent mutations and DNA copy number changes. Our results revealed that the methylome, presenting in early stage samples and associated with the BRAF(V600E) mutation, gradually decreased in the medium and late stages of the disease. An inverse relationship among the other predefined groups and promoter methylation was also revealed except for histologic subtype, whereas the more aggressive, nodular subtype melanomas exhibited hypermethylation as well. The Breslow thickness, which is a continuous variable, allowed for the most precise insight into how promoter methylation decreases from stage to stage. Integrating our methylation results with a high-throughput copy number alteration dataset, local correlations were detected in the MYB and EYA4 genes. With regard to the effects of DNA hypermethylation on melanoma patients' survival, correcting for clinical cofounders, only the KIT gene was associated with a lower overall survival rate. In this study, we demonstrate the strong influence of promoter localized DNA methylation changes on melanoma initiation and show how hypermethylation decreases in melanomas associated with less favourable clinical outcomes. Furthermore, we establish the methylation pattern as part of an integrated apparatus of somatic DNA alterations.
Conflict of interest statement
Figures



Similar articles
-
Marked genetic differences between BRAF and NRAS mutated primary melanomas as revealed by array comparative genomic hybridization.Melanoma Res. 2012 Jun;22(3):202-14. doi: 10.1097/CMR.0b013e328352dbc8. Melanoma Res. 2012. PMID: 22456166
-
The role of CCND1 alterations during the progression of cutaneous malignant melanoma.Tumour Biol. 2012 Dec;33(6):2189-99. doi: 10.1007/s13277-012-0480-6. Epub 2012 Sep 23. Tumour Biol. 2012. PMID: 23001925
-
Aberrant DNA methylation is associated with aggressive clinicopathological features and poor survival in cutaneous melanoma.Br J Dermatol. 2018 Aug;179(2):394-404. doi: 10.1111/bjd.16254. Epub 2018 May 25. Br J Dermatol. 2018. PMID: 29278418
-
BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs.J Clin Pathol. 2005 Jun;58(6):640-4. doi: 10.1136/jcp.2004.022509. J Clin Pathol. 2005. PMID: 15917418 Free PMC article. Review.
-
Aberrant DNA methylation in malignant melanoma.Melanoma Res. 2010 Aug;20(4):253-65. doi: 10.1097/CMR.0b013e328338a35a. Melanoma Res. 2010. PMID: 20418788 Free PMC article. Review.
Cited by
-
BRAFV600E and KIT immunoexpression in early-stage melanoma.An Bras Dermatol. 2019 Oct 17;94(4):458-460. doi: 10.1590/abd1806-4841.20198349. eCollection 2019. An Bras Dermatol. 2019. PMID: 31644622 Free PMC article.
-
Estrogen Receptor β Agonists Differentially Affect the Growth of Human Melanoma Cell Lines.PLoS One. 2015 Jul 30;10(7):e0134396. doi: 10.1371/journal.pone.0134396. eCollection 2015. PLoS One. 2015. PMID: 26225426 Free PMC article.
-
Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities.Clin Epigenetics. 2017 Apr 4;9:34. doi: 10.1186/s13148-017-0332-8. eCollection 2017. Clin Epigenetics. 2017. PMID: 28396701 Free PMC article. Review.
-
HSPB8 counteracts tumor activity of BRAF- and NRAS-mutant melanoma cells by modulation of RAS-prenylation and autophagy.Cell Death Dis. 2022 Nov 18;13(11):973. doi: 10.1038/s41419-022-05365-9. Cell Death Dis. 2022. PMID: 36400750 Free PMC article.
-
Aberrant DNA Methylation Predicts Melanoma-Specific Survival in Patients with Acral Melanoma.Cancers (Basel). 2019 Dec 16;11(12):2031. doi: 10.3390/cancers11122031. Cancers (Basel). 2019. PMID: 31888295 Free PMC article.
References
-
- van den Hurk K, Niessen HE, Veeck J, van den Oord JJ, van Steensel MA, et al. (2012) Genetics and epigenetics of cutaneous malignant melanoma: a concert out of tune. Biochim Biophys Acta 1826: 89–102. - PubMed
-
- Heyn H, Esteller M (2012) DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13: 679–692. - PubMed
-
- Wild L, Flanagan JM (2010) Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta 1806: 50–57. - PubMed
-
- James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, et al. (2003) Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J Nutr 133: 3740S–3747S. - PubMed
-
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, et al. (2013) The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339: 215–218. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

