Genotyping-by-Sequencing in Plants
- PMID: 24832503
- PMCID: PMC4009820
- DOI: 10.3390/biology1030460
Genotyping-by-Sequencing in Plants
Abstract
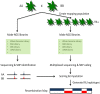
The advent of next-generation DNA sequencing (NGS) technologies has led to the development of rapid genome-wide Single Nucleotide Polymorphism (SNP) detection applications in various plant species. Recent improvements in sequencing throughput combined with an overall decrease in costs per gigabase of sequence is allowing NGS to be applied to not only the evaluation of small subsets of parental inbred lines, but also the mapping and characterization of traits of interest in much larger populations. Such an approach, where sequences are used simultaneously to detect and score SNPs, therefore bypassing the entire marker assay development stage, is known as genotyping-by-sequencing (GBS). This review will summarize the current state of GBS in plants and the promises it holds as a genome-wide genotyping application.
Figures
References
-
- Paran I., Michelmore R.W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 1993;85:985–993. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

