Whole Genome Sequencing and a New Bioinformatics Platform Allow for Rapid Gene Identification in D. melanogaster EMS Screens
- PMID: 24832518
- PMCID: PMC4009818
- DOI: 10.3390/biology1030766
Whole Genome Sequencing and a New Bioinformatics Platform Allow for Rapid Gene Identification in D. melanogaster EMS Screens
Abstract
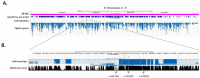
Forward genetic screens in Drosophila melanogaster using ethyl methanesulfonate (EMS) mutagenesis are a powerful approach for identifying genes that modulate specific biological processes in an in vivo setting. The mapping of genes that contain randomly-induced point mutations has become more efficient in Drosophila thanks to the maturation and availability of many types of genetic tools. However, classic approaches to gene mapping are relatively slow and ultimately require extensive Sanger sequencing of candidate chromosomal loci. With the advent of new high-throughput sequencing techniques, it is increasingly efficient to directly re-sequence the whole genome of model organisms. This approach, in combination with traditional chromosomal mapping, has the potential to greatly simplify and accelerate mutation identification in mutants generated in EMS screens. Here we show that next-generation sequencing (NGS) is an accurate and efficient tool for high-throughput sequencing and mutation discovery in Drosophila melanogaster. As a test case, mutant strains of Drosophila that exhibited long-term survival of severed peripheral axons were identified in a forward EMS mutagenesis. All mutants were recessive and fell into a single lethal complementation group, which suggested that a single gene was responsible for the protective axon degenerative phenotype. Whole genome sequencing of these genomes identified the underlying gene ect4. To improve the process of genome wide mutation identification, we developed Genomes Management Application (GEM.app, https://genomics.med.miami.edu), a graphical online user interface to a custom query framework. Using a custom GEM.app query, we were able to identify that each mutant carried a unique non-sense mutation in the gene ect4 (dSarm), which was recently shown by Osterloh et al. to be essential for the activation of axonal degeneration. Our results demonstrate the current advantages and limitations of NGS in Drosophila and we introduce GEM.app as a simple yet powerful genomics analysis tool for the Drosophila community. At a current cost of <$1,000 per genome, NGS should thus become a standard gene discovery tool in EMS induced genetic forward screens.
Figures



References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

