Mutant ELOVL4 that causes autosomal dominant stargardt-3 macular dystrophy is misrouted to rod outer segment disks
- PMID: 24833735
- PMCID: PMC4059079
- DOI: 10.1167/iovs.13-13099
Mutant ELOVL4 that causes autosomal dominant stargardt-3 macular dystrophy is misrouted to rod outer segment disks
Abstract
Purpose: Autosomal dominant Stargardt macular dystrophy caused by mutations in the Elongation of Very Long Chain fatty acids (ELOVL4) gene results in macular degeneration, leading to early childhood blindness. Transgenic mice and pigs expressing mutant ELOVL4 develop progressive photoreceptor degeneration. The mechanism by which these mutations cause macular degeneration remains unclear, but have been hypothesized to involve the loss of an ER-retention dilysine motif located in the extreme C-terminus. Dominant negative mechanisms and reduction in retinal polyunsaturated fatty acids also have been suggested. To understand the molecular mechanisms involved in disease progression in vivo, we addressed the hypothesis that the disease-linked C-terminal truncation mutant of ELOVL4 exerts a dominant negative effect on wild-type (WT) ELOVL4, altering its subcellular localization and function, which subsequently induces retinal degeneration and loss of vision.
Methods: We generated transgenic Xenopus laevis that overexpress HA-tagged murine ELOVL4 variants in rod photoreceptors.
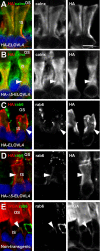
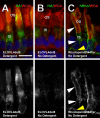
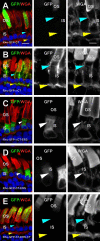
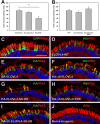
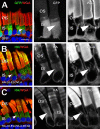
Results: Tagged or untagged WT ELOVL4 localized primarily to inner segments. However, the mutant protein lacking the dilysine motif was mislocalized to post-Golgi compartments and outer segment disks. Coexpression of mutant and WT ELOVL4 in rods did not result in mislocalization of the WT protein to outer segments or in the formation of aggregates. Full-length HA-tagged ELOVL4 lacking the dilysine motif (K308R/K310R) necessary for targeting the WT ELOVL4 protein to the endoplasmic reticulum was similarly mislocalized to outer segments.
Conclusions: We propose that expression and outer segment mislocalization of the disease-linked 5-base-pair deletion mutant ELOVL4 protein alters photoreceptor structure and function, which subsequently results in retinal degeneration, and suggest three possible mechanisms by which mutant ELOVL4 may induce retinal degeneration in STGD3.
Keywords: autosomal dominant Stargardt-like macular dystrophy (STGD3); elongation of very long chain fatty acids-4 (ELOVL4); photoreceptor outer segment; retinal degeneration.
Copyright 2014 The Association for Research in Vision and Ophthalmology, Inc.
Figures



Similar articles
-
Early Onset Ultrastructural and Functional Defects in RPE and Photoreceptors of a Stargardt-Like Macular Dystrophy (STGD3) Transgenic Mouse Model.Invest Ophthalmol Vis Sci. 2015 Nov;56(12):7109-21. doi: 10.1167/iovs.15-17567. Invest Ophthalmol Vis Sci. 2015. PMID: 26529045
-
Role of ELOVL4 and very long-chain polyunsaturated fatty acids in mouse models of Stargardt type 3 retinal degeneration.Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):5181-6. doi: 10.1073/pnas.1214707110. Epub 2013 Mar 11. Proc Natl Acad Sci U S A. 2013. PMID: 23479632 Free PMC article.
-
Deciphering mutant ELOVL4 activity in autosomal-dominant Stargardt macular dystrophy.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5446-51. doi: 10.1073/pnas.1217251110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509295 Free PMC article.
-
Genetics and molecular pathology of Stargardt-like macular degeneration.Prog Retin Eye Res. 2010 May;29(3):191-207. doi: 10.1016/j.preteyeres.2010.01.001. Epub 2010 Jan 21. Prog Retin Eye Res. 2010. PMID: 20096366 Free PMC article. Review.
-
Dominant Stargardt Macular Dystrophy (STGD3) and ELOVL4.Adv Exp Med Biol. 2014;801:447-53. doi: 10.1007/978-1-4614-3209-8_57. Adv Exp Med Biol. 2014. PMID: 24664730 Review.
Cited by
-
Adiponectin receptor 1 conserves docosahexaenoic acid and promotes photoreceptor cell survival.Nat Commun. 2015 Mar 4;6:6228. doi: 10.1038/ncomms7228. Nat Commun. 2015. PMID: 25736573 Free PMC article.
-
Defective phagosome motility and degradation in cell nonautonomous RPE pathogenesis of a dominant macular degeneration.Proc Natl Acad Sci U S A. 2018 May 22;115(21):5468-5473. doi: 10.1073/pnas.1709211115. Epub 2018 May 7. Proc Natl Acad Sci U S A. 2018. PMID: 29735674 Free PMC article.
-
Photoreceptor-induced RPE phagolysosomal maturation defects in Stargardt-like Maculopathy (STGD3).Sci Rep. 2018 Apr 13;8(1):5944. doi: 10.1038/s41598-018-24357-4. Sci Rep. 2018. PMID: 29654292 Free PMC article.
-
Updates on Emerging Interventions for Autosomal Recessive ABCA4-Associated Stargardt Disease.J Clin Med. 2023 Sep 27;12(19):6229. doi: 10.3390/jcm12196229. J Clin Med. 2023. PMID: 37834872 Free PMC article. Review.
-
Elovl4 5-bp deletion does not accelerate cone photoreceptor degeneration in an all-cone mouse.PLoS One. 2018 Jan 2;13(1):e0190514. doi: 10.1371/journal.pone.0190514. eCollection 2018. PLoS One. 2018. PMID: 29293603 Free PMC article.
References
-
- Zhang K, Kniazeva M, Han M, et al. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet. 2001; 27: 89–93 - PubMed
-
- Carmona-Antonanzas G, Monroig O, Dick JR, Davie A, Tocher DR. Biosynthesis of very long-chain fatty acids (C>24) in Atlantic salmon: cloning, functional characterisation, and tissue distribution of an Elovl4 elongase. Comp Biochem Physiol B Biochem Mol Biol. 2011; 159: 122–129 - PubMed
-
- Ambasudhan R, Wang X, Jablonski MM, et al. Atrophic macular degeneration mutations in ELOVL4 result in the intracellular misrouting of the protein. Genomics. 2004; 83: 615–625 - PubMed
-
- Karan G, Yang Z, Zhang K. Expression of wild type and mutant ELOVL4 in cell culture: subcellular localization and cell viability. Mol Vis. 2004; 10: 248–253 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

