A convenient, optimized pipeline for isolation, fluorescence microscopy and molecular analysis of live single cells
- PMID: 24834016
- PMCID: PMC4022543
- DOI: 10.1186/1480-9222-16-9
A convenient, optimized pipeline for isolation, fluorescence microscopy and molecular analysis of live single cells
Abstract
Background: Heterogeneity within cell populations is relevant to the onset and progression of disease, as well as development and maintenance of homeostasis. Analysis and understanding of the roles of heterogeneity in biological systems require methods and technologies that are capable of single cell resolution. Single cell gene expression analysis by RT-qPCR is an established technique for identifying transcriptomic heterogeneity in cellular populations, but it generally requires specialized equipment or tedious manipulations for cell isolation.
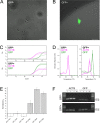
Results: We describe the optimization of a simple, inexpensive and rapid pipeline which includes isolation and culture of live single cells as well as fluorescence microscopy and gene expression analysis of the same single cells by RT-qPCR. We characterize the efficiency of single cell isolation and demonstrate our method by identifying single GFP-expressing cells from a mixed population of GFP-positive and negative cells by correlating fluorescence microscopy and RT-qPCR.
Conclusions: Single cell gene expression analysis by RT-qPCR is a convenient means for investigating cellular heterogeneity, but is most useful when correlating observations with additional measurements. We demonstrate a convenient and simple pipeline for multiplexing single cell RT-qPCR with fluorescence microscopy which is adaptable to other molecular analyses.
Keywords: Fluorescence microscopy; Gene expression analysis; RT-qPCR; Single cell.
Figures


References
-
- De Souza N. Single-cell methods. Nat Methods. 2011;9:35.
LinkOut - more resources
Full Text Sources
Other Literature Sources

