Modelization of the regulation of protein synthesis following fertilization in sea urchin shows requirement of two processes: a destabilization of eIF4E:4E-BP complex and a great stimulation of the 4E-BP-degradation mechanism, both rapamycin-sensitive
- PMID: 24834072
- PMCID: PMC4018528
- DOI: 10.3389/fgene.2014.00117
Modelization of the regulation of protein synthesis following fertilization in sea urchin shows requirement of two processes: a destabilization of eIF4E:4E-BP complex and a great stimulation of the 4E-BP-degradation mechanism, both rapamycin-sensitive
Abstract
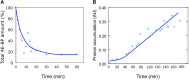
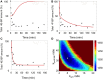
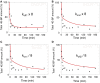
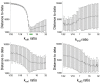
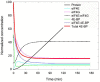
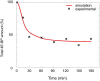
Fertilization of sea urchin eggs involves an increase in protein synthesis associated with a decrease in the amount of the translation initiation inhibitor 4E-BP. A highly simple reaction model for the regulation of protein synthesis was built and was used to simulate the physiological changes in the total 4E-BP amount observed during time after fertilization. Our study evidenced that two changes occurring at fertilization are necessary to fit with experimental data. The first change was an 8-fold increase in the dissociation parameter (koff1) of the eIF4E:4E-BP complex. The second was an important 32.5-fold activation of the degradation mechanism of the protein 4E-BP. Additionally, the changes in both processes should occur in 5 min time interval post-fertilization. To validate the model, we checked that the kinetic of the predicted 4.2-fold increase of eIF4E:eIF4G complex concentration at fertilization matched the increase of protein synthesis experimentally observed after fertilization (6.6-fold, SD = 2.3, n = 8). The minimal model was also used to simulate changes observed after fertilization in the presence of rapamycin, a FRAP/mTOR inhibitor. The model showed that the eIF4E:4E-BP complex destabilization was impacted and surprisingly, that the mechanism of 4E-BP degradation was also strongly affected, therefore suggesting that both processes are controlled by the protein kinase FRAP/mTOR.
Keywords: deterministic model; mechanisms of fertilization; sea urchin embryos; translation simulation; translational control.
Figures


References
-
- Abiko F., Tomoo K., Mizuno A., Morino S., Imataka H., Ishida T. (2007). Binding preference of eIF4E for 4E-binding protein isoform and function of eIF4E N-terminal flexible region for interaction, studied by SPR analysis. Biochem. Biophys. Res. Commun. 355, 667–672 10.1016/j.bbrc.2007.01.198 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

