A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus
- PMID: 24835457
- PMCID: PMC4046227
- DOI: 10.1016/j.cub.2014.04.009
A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus
Abstract
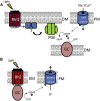
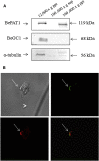
Sensing light is the fundamental property of visual systems, with vision in animals being based almost exclusively on opsin photopigments [1]. Rhodopsin also acts as a photoreceptor linked to phototaxis in green algae [2, 3] and has been implicated by chemical means as a light sensor in the flagellated swimming zoospores of the fungus Allomyces reticulatus [4]; however, the signaling mechanism in these fungi remains unknown. Here we use a combination of genome sequencing and molecular inhibition experiments with light-sensing phenotype studies to examine the signaling pathway involved in visual perception in the closely related fungus Blastocladiella emersonii. Our data show that in these fungi, light perception is accomplished by the function of a novel gene fusion (BeGC1) of a type I (microbial) rhodopsin domain and guanylyl cyclase catalytic domain. Photobleaching of rhodopsin function prevents accumulation of cGMP levels and phototaxis of fungal zoospores exposed to green light, whereas inhibition of guanylyl cyclase activity negatively affects fungal phototaxis. Immunofluorescence microscopy localizes the BeGC1 protein to the external surface of the zoospore eyespot positioned close to the base of the swimming flagellum [4, 5], demonstrating this is a photoreceptive organelle composed of lipid droplets. Taken together, these data indicate that Blastocladiomycota fungi have a cGMP signaling pathway involved in phototaxis similar to the vertebrate vision-signaling cascade but composed of protein domain components arranged as a novel gene fusion architecture and of distant evolutionary ancestry to type II rhodopsins of animals.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Foster K.W., Saranak J., Patel N., Zarilli G., Okabe M., Kline T., Nakanishi K. A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature. 1984;311:756–759. - PubMed
-
- Saranak J., Foster K.W. Rhodopsin guides fungal phototaxis. Nature. 1997;387:465–466. - PubMed
-
- Cantino E.C., Truesdell L.C. Organization and fine structure of the side body and its lipid sac in the zoospore of Blastocladiella emersonii. Mycologia. 1970;62:548–567. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

