Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary
- PMID: 24835588
- PMCID: PMC4120086
- DOI: 10.1101/gr.168997.113
Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary
Abstract
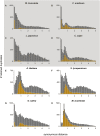
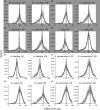
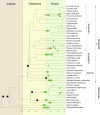
Ancient whole-genome duplications (WGDs), also referred to as paleopolyploidizations, have been reported in most evolutionary lineages. Their attributed role remains a major topic of discussion, ranging from an evolutionary dead end to a road toward evolutionary success, with evidence supporting both fates. Previously, based on dating WGDs in a limited number of plant species, we found a clustering of angiosperm paleopolyploidizations around the Cretaceous-Paleogene (K-Pg) extinction event about 66 million years ago. Here we revisit this finding, which has proven controversial, by combining genome sequence information for many more plant lineages and using more sophisticated analyses. We include 38 full genome sequences and three transcriptome assemblies in a Bayesian evolutionary analysis framework that incorporates uncorrelated relaxed clock methods and fossil uncertainty. In accordance with earlier findings, we demonstrate a strongly nonrandom pattern of genome duplications over time with many WGDs clustering around the K-Pg boundary. We interpret these results in the context of recent studies on invasive polyploid plant species, and suggest that polyploid establishment is promoted during times of environmental stress. We argue that considering the evolutionary potential of polyploids in light of the environmental and ecological conditions present around the time of polyploidization could mitigate the stark contrast in the proposed evolutionary fates of polyploids.
© 2014 Vanneste et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Abbasi AA 2010. Piecemeal or big bangs: correlating the vertebrate evolution with proposed models of gene expansion events. Nat Rev Genet 11: 166. - PubMed
-
- Akaike H 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19: 716–723
-
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444: 171–178 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
