Digital expression profiling identifies RUNX2, CDC5L, MDM2, RECQL4, and CDK4 as potential predictive biomarkers for neo-adjuvant chemotherapy response in paediatric osteosarcoma
- PMID: 24835790
- PMCID: PMC4023931
- DOI: 10.1371/journal.pone.0095843
Digital expression profiling identifies RUNX2, CDC5L, MDM2, RECQL4, and CDK4 as potential predictive biomarkers for neo-adjuvant chemotherapy response in paediatric osteosarcoma
Abstract
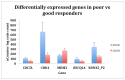
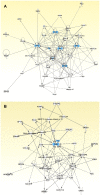
Osteosarcoma is the most common malignancy of bone, and occurs most frequently in children and adolescents. Currently, the most reliable technique for determining a patients' prognosis is measurement of histopathologic tumor necrosis following pre-operative neo-adjuvant chemotherapy. Unfavourable prognosis is indicated by less than 90% estimated necrosis of the tumor. Neither genetic testing nor molecular biomarkers for diagnosis and prognosis have been described for osteosarcomas. We used the novel nanoString mRNA digital expression analysis system to analyse gene expression in 32 patients with sporadic paediatric osteosarcoma. This system used specific molecular barcodes to quantify expression of a set of 17 genes associated with osteosarcoma tumorigenesis. Five genes, from this panel, which encoded the bone differentiation regulator RUNX2, the cell cycle regulator CDC5L, the TP53 transcriptional inactivator MDM2, the DNA helicase RECQL4, and the cyclin-dependent kinase gene CDK4, were differentially expressed in tumors that responded poorly to neo-adjuvant chemotherapy. Analysis of the signalling relationships of these genes, as well as other expression markers of osteosarcoma, indicated that gene networks linked to RB1, TP53, PI3K, PTEN/Akt, myc and RECQL4 are associated with osteosarcoma. The discovery of these networks provides a basis for further experimental studies of role of the five genes (RUNX2, CDC5L, MDM2, RECQL4, and CDK4) in differential response to chemotherapy.
Conflict of interest statement
Figures

References
-
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, et al. (2002) Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 20(3): 776–790. - PubMed
-
- Selvarajah S, Yoshimoto M, Ludkovski O, Park PC, Bayani J, et al. (2008) Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenet Genome Res 122(1): 5–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

