AKAP-anchored PKA maintains neuronal L-type calcium channel activity and NFAT transcriptional signaling
- PMID: 24835999
- PMCID: PMC4136445
- DOI: 10.1016/j.celrep.2014.04.027
AKAP-anchored PKA maintains neuronal L-type calcium channel activity and NFAT transcriptional signaling
Abstract
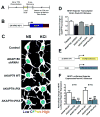
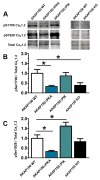
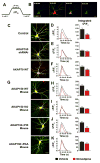
L-type voltage-gated Ca2+ channels (LTCC) couple neuronal excitation to gene transcription. LTCC activity is elevated by the cyclic AMP (cAMP)-dependent protein kinase (PKA) and depressed by the Ca2+-dependent phosphatase calcineurin (CaN), and both enzymes are localized to the channel by A-kinase anchoring protein 79/150 (AKAP79/150). AKAP79/150 anchoring of CaN also promotes LTCC activation of transcription through dephosphorylation of the nuclear factor of activated T cells (NFAT). We report here that the basal activity of AKAP79/150-anchored PKA maintains neuronal LTCC coupling to CaN-NFAT signaling by preserving LTCC phosphorylation in opposition to anchored CaN. Genetic disruption of AKAP-PKA anchoring promoted redistribution of the kinase out of postsynaptic dendritic spines, profound decreases in LTCC phosphorylation and Ca2+ influx, and impaired NFAT movement to the nucleus and activation of transcription. Thus, LTCC-NFAT transcriptional signaling in neurons requires precise organization and balancing of PKA and CaN activities in the channel nanoenvironment, which is only made possible by AKAP79/150 scaffolding.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
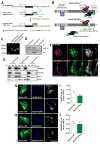
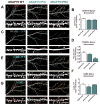
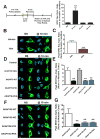
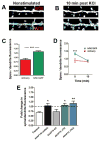
Comment in
-
AKAP5 keeps L-type channels and NFAT on their toes.Cell Rep. 2014 Jun 12;7(5):1341-1342. doi: 10.1016/j.celrep.2014.05.052. Cell Rep. 2014. PMID: 24926793 Free PMC article.
References
-
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. - PubMed
-
- Belfield JL, Whittaker C, Cader MZ, Chawla S. Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J Biol Chem. 2006;281:27724–27732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1TR001082/TR/NCATS NIH HHS/United States
- T32 NS099042/NS/NINDS NIH HHS/United States
- R56 NS040701/NS/NINDS NIH HHS/United States
- R01NS040701/NS/NINDS NIH HHS/United States
- R01HL085372/HL/NHLBI NIH HHS/United States
- R01 MH080291/MH/NIMH NIH HHS/United States
- R01 HL085372/HL/NHLBI NIH HHS/United States
- P30NS048154/NS/NINDS NIH HHS/United States
- R01GM48231/GM/NIGMS NIH HHS/United States
- R01 MH102338/MH/NIMH NIH HHS/United States
- P30 NS048154/NS/NINDS NIH HHS/United States
- R01MH080291/MH/NIMH NIH HHS/United States
- R01 GM048231/GM/NIGMS NIH HHS/United States
- R01MH102338/MH/NIMH NIH HHS/United States
- R01 NS040701/NS/NINDS NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

