Contrasting X-linked and autosomal diversity across 14 human populations
- PMID: 24836452
- PMCID: PMC4121480
- DOI: 10.1016/j.ajhg.2014.04.011
Contrasting X-linked and autosomal diversity across 14 human populations
Abstract
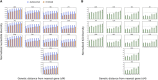
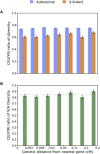
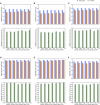
Contrasting the genetic diversity of the human X chromosome (X) and autosomes has facilitated understanding historical differences between males and females and the influence of natural selection. Previous studies based on smaller data sets have left questions regarding how empirical patterns extend to additional populations and which forces can explain them. Here, we address these questions by analyzing the ratio of X-to-autosomal (X/A) nucleotide diversity with the complete genomes of 569 females from 14 populations. Results show that X/A diversity is similar within each continental group but notably lower in European (EUR) and East Asian (ASN) populations than in African (AFR) populations. X/A diversity increases in all populations with increasing distance from genes, highlighting the stronger impact of diversity-reducing selection on X than on the autosomes. However, relative X/A diversity (between two populations) is invariant with distance from genes, suggesting that selection does not drive the relative reduction in X/A diversity in non-Africans (0.842 ± 0.012 for EUR-to-AFR and 0.820 ± 0.032 for ASN-to-AFR comparisons). Finally, an array of models with varying population bottlenecks, expansions, and migration from the latest studies of human demographic history account for about half of the observed reduction in relative X/A diversity from the expected value of 1. They predict values between 0.91 and 0.94 for EUR-to-AFR comparisons and between 0.91 and 0.92 for ASN-to-AFR comparisons. Further reductions can be predicted by more extreme demographic events in excess of those captured by the latest studies but, in the absence of these, also by historical sex-biased demographic events or other processes.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

