Bayesian approach to single-cell differential expression analysis
- PMID: 24836921
- PMCID: PMC4112276
- DOI: 10.1038/nmeth.2967
Bayesian approach to single-cell differential expression analysis
Abstract
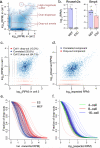
Single-cell data provide a means to dissect the composition of complex tissues and specialized cellular environments. However, the analysis of such measurements is complicated by high levels of technical noise and intrinsic biological variability. We describe a probabilistic model of expression-magnitude distortions typical of single-cell RNA-sequencing measurements, which enables detection of differential expression signatures and identification of subpopulations of cells in a way that is more tolerant of noise.
Figures
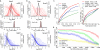
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

