Dynamics of expression and maturation of the type III secretion system of enteropathogenic Escherichia coli
- PMID: 24837293
- PMCID: PMC4135678
- DOI: 10.1128/JB.00069-14
Dynamics of expression and maturation of the type III secretion system of enteropathogenic Escherichia coli
Abstract
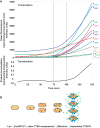
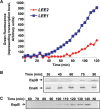
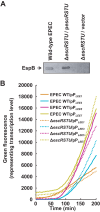
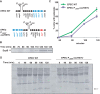
Enteropathogenic Escherichia coli (EPEC) is a major cause of food poisoning, leading to significant morbidity and mortality. EPEC virulence is dependent on a type III secretion system (T3SS), a molecular syringe employed by EPEC to inject effector proteins into host cells. The injected effector proteins subvert host cellular functions to the benefit of the infecting bacteria. The T3SS and related genes reside in several operons clustered in the locus of enterocyte effacement (LEE). We carried out simultaneous analysis of the expression dynamics of all the LEE promoters and the rate of maturation of the T3SS. The results showed that expression of the LEE1 operon is activated immediately upon shifting the culture to inducing conditions, while expression of other LEE promoters is activated only ∼70 min postinduction. Parallel analysis showed that the T3SS becomes functional around 100 min postinduction. The T3SS core proteins EscS, EscT, EscU, and EscR are predicted to be involved in the first step of T3SS assembly and are therefore included among the LEE1 genes. However, interfering with the temporal regulation of EscS, EscT, EscU, and EscR expression has only a marginal effect on the rate of the T3SS assembly. This study provides a comprehensive description of the transcription dynamics of all the LEE genes and correlates it to that of T3SS biogenesis.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

