Gene discovery for enzymes involved in limonene modification or utilization by the mountain pine beetle-associated pathogen Grosmannia clavigera
- PMID: 24837377
- PMCID: PMC4148792
- DOI: 10.1128/AEM.00670-14
Gene discovery for enzymes involved in limonene modification or utilization by the mountain pine beetle-associated pathogen Grosmannia clavigera
Abstract
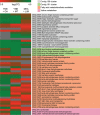
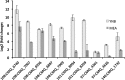
To successfully colonize and eventually kill pine trees, Grosmannia clavigera (Gs cryptic species), the main fungal pathogen associated with the mountain pine beetle (Dendroctonus ponderosae), has developed multiple mechanisms to overcome host tree chemical defenses, of which terpenoids are a major component. In addition to a monoterpene efflux system mediated by a recently discovered ABC transporter, Gs has genes that are highly induced by monoterpenes and that encode enzymes that modify or utilize monoterpenes [especially (+)-limonene]. We showed that pine-inhabiting Ophiostomale fungi are tolerant to monoterpenes, but only a few, including Gs, are known to utilize monoterpenes as a carbon source. Gas chromatography-mass spectrometry (GC-MS) revealed that Gs can modify (+)-limonene through various oxygenation pathways, producing carvone, p-mentha-2,8-dienol, perillyl alcohol, and isopiperitenol. It can also degrade (+)-limonene through the C-1-oxygenated pathway, producing limonene-1,2-diol as the most abundant intermediate. Transcriptome sequencing (RNA-seq) data indicated that Gs may utilize limonene 1,2-diol through beta-oxidation and then valine and tricarboxylic acid (TCA) metabolic pathways. The data also suggested that at least two gene clusters, located in genome contigs 108 and 161, were highly induced by monoterpenes and may be involved in monoterpene degradation processes. Further, gene knockouts indicated that limonene degradation required two distinct Baeyer-Villiger monooxygenases (BVMOs), an epoxide hydrolase and an enoyl coenzyme A (enoyl-CoA) hydratase. Our work provides information on enzyme-mediated limonene utilization or modification and a more comprehensive understanding of the interaction between an economically important fungal pathogen and its host's defense chemicals.
Figures



Similar articles
-
A specialized ABC efflux transporter GcABC-G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle-associated fungal pathogen of pine trees.New Phytol. 2013 Feb;197(3):886-898. doi: 10.1111/nph.12063. Epub 2012 Dec 17. New Phytol. 2013. PMID: 23252416
-
Influence of water deficit on the molecular responses of Pinus contorta × Pinus banksiana mature trees to infection by the mountain pine beetle fungal associate, Grosmannia clavigera.Tree Physiol. 2014 Nov;34(11):1220-39. doi: 10.1093/treephys/tpt101. Epub 2013 Dec 5. Tree Physiol. 2014. PMID: 24319029 Free PMC article.
-
The cytochromes P450 of Grosmannia clavigera: Genome organization, phylogeny, and expression in response to pine host chemicals.Fungal Genet Biol. 2013 Jan;50:72-81. doi: 10.1016/j.fgb.2012.10.002. Epub 2012 Oct 27. Fungal Genet Biol. 2013. PMID: 23111002
-
Mountain Pine Beetle Epidemic: An Interplay of Terpenoids in Host Defense and Insect Pheromones.Annu Rev Plant Biol. 2022 May 20;73:475-494. doi: 10.1146/annurev-arplant-070921-103617. Epub 2022 Feb 7. Annu Rev Plant Biol. 2022. PMID: 35130442 Review.
-
Antitumorigenic effects of limonene and perillyl alcohol against pancreatic and breast cancer.Adv Exp Med Biol. 1996;401:131-6. doi: 10.1007/978-1-4613-0399-2_10. Adv Exp Med Biol. 1996. PMID: 8886131 Review.
Cited by
-
Bark Beetles Utilize Ophiostomatoid Fungi to Circumvent Host Tree Defenses.Metabolites. 2023 Feb 6;13(2):239. doi: 10.3390/metabo13020239. Metabolites. 2023. PMID: 36837858 Free PMC article.
-
Monoterpene biotransformation by Colletotrichum species.Biotechnol Lett. 2018 Mar;40(3):561-567. doi: 10.1007/s10529-017-2503-2. Epub 2017 Dec 29. Biotechnol Lett. 2018. PMID: 29288353
-
Endophyte genomes support greater metabolic gene cluster diversity compared with non-endophytes in Trichoderma.PLoS One. 2023 Dec 21;18(12):e0289280. doi: 10.1371/journal.pone.0289280. eCollection 2023. PLoS One. 2023. PMID: 38127903 Free PMC article.
-
Specialized plant biochemistry drives gene clustering in fungi.ISME J. 2018 Jun;12(7):1694-1705. doi: 10.1038/s41396-018-0075-3. Epub 2018 Feb 20. ISME J. 2018. PMID: 29463891 Free PMC article.
-
Identification of Fungal Limonene-3-Hydroxylase for Biotechnological Menthol Production.Appl Environ Microbiol. 2021 Apr 27;87(10):e02873-20. doi: 10.1128/AEM.02873-20. Print 2021 Apr 27. Appl Environ Microbiol. 2021. PMID: 33637576 Free PMC article.
References
-
- Safranyik L, Carroll AL, Regniere J, Langor DW, Riel WG, Shore TL, Peter B, Cooke BJ, Nealis VG, Taylor SW. 2010. Potential for range expansion of mountain pine beetle into the boreal forest of North America. Can. Entomol. 142:415–442. 10.4039/n08-CPA01 - DOI
-
- de la Giroday HC, Carroll AL, Aukema BH. 2012. Breach of the northern Rocky Mountain geoclimatic barrier: initiation of range expansion by the mountain pine beetle. J. Biogeogr. 39:1112–1123. 10.1111/j.1365-2699.2011.02673.x - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

