Characterization of a nitrilase and a nitrile hydratase from Pseudomonas sp. strain UW4 that converts indole-3-acetonitrile to indole-3-acetic acid
- PMID: 24837382
- PMCID: PMC4148799
- DOI: 10.1128/AEM.00649-14
Characterization of a nitrilase and a nitrile hydratase from Pseudomonas sp. strain UW4 that converts indole-3-acetonitrile to indole-3-acetic acid
Abstract
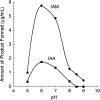
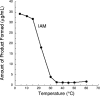
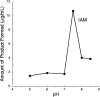
Indole-3-acetic acid (IAA) is a fundamental phytohormone with the ability to control many aspects of plant growth and development. Pseudomonas sp. strain UW4 is a rhizospheric plant growth-promoting bacterium that produces and secretes IAA. While several putative IAA biosynthetic genes have been reported in this bacterium, the pathways leading to the production of IAA in strain UW4 are unclear. Here, the presence of the indole-3-acetamide (IAM) and indole-3-acetaldoxime/indole-3-acetonitrile (IAOx/IAN) pathways of IAA biosynthesis is described, and the specific role of two of the enzymes (nitrilase and nitrile hydratase) that mediate these pathways is assessed. The genes encoding these two enzymes were expressed in Escherichia coli, and the enzymes were isolated and characterized. Substrate-feeding assays indicate that the nitrilase produces both IAM and IAA from the IAN substrate, while the nitrile hydratase only produces IAM. The two nitrile-hydrolyzing enzymes have very different temperature and pH optimums. Nitrilase prefers a temperature of 50°C and a pH of 6, while nitrile hydratase prefers 4°C and a pH of 7.5. Based on multiple sequence alignments and motif analyses, physicochemical properties and enzyme assays, it is concluded that the UW4 nitrilase has an aromatic substrate specificity. The nitrile hydratase is identified as an iron-type metalloenzyme that does not require the help of a P47K activator protein to be active. These data are interpreted in terms of a preliminary model for the biosynthesis of IAA in this bacterium.
Figures

Similar articles
-
The Plant Growth-Promoting Rhizobacterium Variovorax boronicumulans CGMCC 4969 Regulates the Level of Indole-3-Acetic Acid Synthesized from Indole-3-Acetonitrile.Appl Environ Microbiol. 2018 Aug 1;84(16):e00298-18. doi: 10.1128/AEM.00298-18. Print 2018 Aug 15. Appl Environ Microbiol. 2018. PMID: 29884755 Free PMC article.
-
The Nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid.Plant Physiol. 2003 Oct;133(2):794-802. doi: 10.1104/pp.103.026609. Epub 2003 Sep 4. Plant Physiol. 2003. PMID: 12972653 Free PMC article.
-
Bioconversion of indole-3-acetonitrile by the N2-fixing bacterium Ensifer meliloti CGMCC 7333 and its Escherichia coli-expressed nitrile hydratase.Int Microbiol. 2020 May;23(2):225-232. doi: 10.1007/s10123-019-00094-0. Epub 2019 Aug 13. Int Microbiol. 2020. PMID: 31410668
-
Biochemistry and biotechnology of mesophilic and thermophilic nitrile metabolizing enzymes.Extremophiles. 1998 Aug;2(3):207-16. doi: 10.1007/s007920050062. Extremophiles. 1998. PMID: 9783167 Review.
-
Microbial transformation of nitriles to high-value acids or amides.Adv Biochem Eng Biotechnol. 2009;113:33-77. doi: 10.1007/10_2008_25. Adv Biochem Eng Biotechnol. 2009. PMID: 19475377 Review.
Cited by
-
Identification and Characterization of Auxin/IAA Biosynthesis Pathway in the Rice Blast Fungus Magnaporthe oryzae.J Fungi (Basel). 2022 Feb 21;8(2):208. doi: 10.3390/jof8020208. J Fungi (Basel). 2022. PMID: 35205962 Free PMC article.
-
Screening and characterization of a nitrilase with significant nitrile hydratase activity.Biotechnol Lett. 2022 Oct;44(10):1163-1173. doi: 10.1007/s10529-022-03291-6. Epub 2022 Sep 1. Biotechnol Lett. 2022. PMID: 36050605
-
Plant growth-promoting effect and genomic analysis of the P. putida LWPZF isolated from C. japonicum rhizosphere.AMB Express. 2022 Aug 2;12(1):101. doi: 10.1186/s13568-022-01445-3. AMB Express. 2022. PMID: 35917000 Free PMC article.
-
Identification and combinatorial engineering of indole-3-acetic acid synthetic pathways in Paenibacillus polymyxa.Biotechnol Biofuels Bioprod. 2022 Aug 11;15(1):81. doi: 10.1186/s13068-022-02181-3. Biotechnol Biofuels Bioprod. 2022. PMID: 35953838 Free PMC article.
-
Employing Genomic Tools to Explore the Molecular Mechanisms behind the Enhancement of Plant Growth and Stress Resilience Facilitated by a Burkholderia Rhizobacterial Strain.Int J Mol Sci. 2024 May 31;25(11):6091. doi: 10.3390/ijms25116091. Int J Mol Sci. 2024. PMID: 38892282 Free PMC article.
References
-
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. 2011. Vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23:550–566. 10.1105/tpc.110.075267 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

