SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18
- PMID: 24837546
- PMCID: PMC4060006
- DOI: 10.7554/eLife.02272
SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18
Abstract
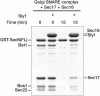
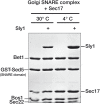
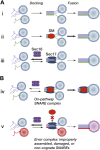
Secretory and endolysosomal fusion events are driven by SNAREs and cofactors, including Sec17/α-SNAP, Sec18/NSF, and Sec1/Munc18 (SM) proteins. SMs are essential for fusion in vivo, but the basis of this requirement is enigmatic. We now report that, in addition to their established roles as fusion accelerators, SM proteins Sly1 and Vps33 directly shield SNARE complexes from Sec17- and Sec18-mediated disassembly. In vivo, wild-type Sly1 and Vps33 function are required to withstand overproduction of Sec17. In vitro, Sly1 and Vps33 impede SNARE complex disassembly by Sec18 and ATP. Unexpectedly, Sec17 directly promotes selective loading of Sly1 and Vps33 onto cognate SNARE complexes. A large thermodynamic barrier limits SM binding, implying that significant conformational rearrangements are involved. In a working model, Sec17 and SMs accelerate fusion mediated by cognate SNARE complexes and protect them from NSF-mediated disassembly, while mis-assembled or non-cognate SNARE complexes are eliminated through kinetic proofreading by Sec18.DOI: http://dx.doi.org/10.7554/eLife.02272.001.
Keywords: Golgi; HOPS; SNARE; docking; lysosome; membrane.
Copyright © 2014, Lobingier et al.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures













Comment in
-
To protect or reject.Elife. 2014 Jun 17;3:e03374. doi: 10.7554/eLife.03374. Elife. 2014. PMID: 24940001 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

