CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity
- PMID: 24837675
- PMCID: PMC4105207
- DOI: 10.1016/j.molcel.2014.04.012
CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity
Abstract
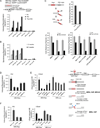
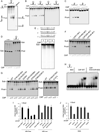
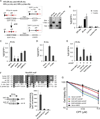
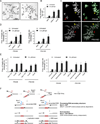
Chromosomal rearrangements often occur at genomic loci with DNA secondary structures, such as common fragile sites (CFSs) and palindromic repeats. We developed assays in mammalian cells that revealed CFS-derived AT-rich sequences and inverted Alu repeats (Alu-IRs) are mitotic recombination hotspots, requiring the repair functions of carboxy-terminal binding protein (CtBP)-interacting protein (CtIP) and the Mre11/Rad50/Nbs1 complex (MRN). We also identified an endonuclease activity of CtIP that is dispensable for end resection and homologous recombination (HR) at I-SceI-generated "clean" double-strand breaks (DSBs) but is required for repair of DSBs occurring at CFS-derived AT-rich sequences. In addition, CtIP nuclease-defective mutants are impaired in Alu-IRs-induced mitotic recombination. These studies suggest that an end resection-independent CtIP function is important for processing DSB ends with secondary structures to promote HR. Furthermore, our studies uncover an important role of MRN, CtIP, and their associated nuclease activities in protecting CFSs in mammalian cells.
Copyright © 2014 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Abeysinghe SS, Stenson PD, Krawczak M, Cooper DN. Gross Rearrangement Breakpoint Database (GRaBD) Human mutation. 2004;23:219–221. - PubMed
-
- Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 2006;5:1126–1135. - PubMed
-
- Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nature biotechnology. 2003;21:321–324. - PubMed
-
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. - PubMed
-
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Seminars in cancer biology. 2010;20:222–233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

