Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection
- PMID: 24837676
- PMCID: PMC4079050
- DOI: 10.1016/j.molcel.2014.04.011
Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection
Abstract
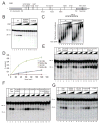
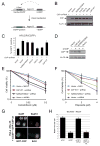
The carboxy-terminal binding protein (CtBP)-interacting protein (CtIP) is known to function in 5' strand resection during homologous recombination, similar to the budding yeast Sae2 protein, but its role in this process is unclear. Here, we characterize recombinant human CtIP and find that it exhibits 5' flap endonuclease activity on branched DNA structures, independent of the MRN complex. Phosphorylation of CtIP at known damage-dependent sites and other sites is essential for its catalytic activity, although the S327 and T847 phosphorylation sites are dispensable. A catalytic mutant of CtIP that is deficient in endonuclease activity exhibits wild-type levels of homologous recombination at restriction enzyme-generated breaks but is deficient in processing topoisomerase adducts and radiation-induced breaks in human cells, suggesting that the nuclease activity of CtIP is specifically required for the removal of DNA adducts at sites of DNA breaks.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

