CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains
- PMID: 24838564
- PMCID: PMC4086077
- DOI: 10.1093/nar/gku359
CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains
Abstract
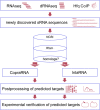
CopraRNA (Comparative prediction algorithm for small RNA targets) is the most recent asset to the Freiburg RNA Tools webserver. It incorporates and extends the functionality of the existing tool IntaRNA (Interacting RNAs) in order to predict targets, interaction domains and consequently the regulatory networks of bacterial small RNA molecules. The CopraRNA prediction results are accompanied by extensive postprocessing methods such as functional enrichment analysis and visualization of interacting regions. Here, we introduce the functionality of the CopraRNA and IntaRNA webservers and give detailed explanations on their postprocessing functionalities. Both tools are freely accessible at http://rna.informatik.uni-freiburg.de.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Mitschke J., Georg J., Scholz I., Sharma C.M., Dienst D., Bantscheff J., Voß B., Steglich C., Wilde A., Vogel J., et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2124–2129. - PMC - PubMed
-
- Georg J., Hess W.R. Regulatory RNAs in cyanobacteria: developmental decisions, stress responses and a plethora of chromosomally encoded cis-antisense RNAs. Biol. Chem. 2011;392:291–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

