Aminopeptidase N1 (EtAPN1), an M1 metalloprotease of the apicomplexan parasite Eimeria tenella, participates in parasite development
- PMID: 24839124
- PMCID: PMC4135734
- DOI: 10.1128/EC.00062-14
Aminopeptidase N1 (EtAPN1), an M1 metalloprotease of the apicomplexan parasite Eimeria tenella, participates in parasite development
Abstract
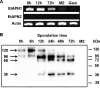
Aminopeptidases N are metalloproteases of the M1 family that have been reported in numerous apicomplexan parasites, including Plasmodium, Toxoplasma, Cryptosporidium, and Eimeria. While investigating the potency of aminopeptidases as therapeutic targets against coccidiosis, one of the most important avian diseases caused by the genus Eimeria, we identified and characterized Eimeria tenella aminopeptidase N1 (EtAPN1). Its inhibition by bestatin and amastatin, as well as its reactivation by divalent ions, is typical of zinc-dependent metalloproteases. EtAPN1 shared a similar sequence, three-dimensional structure, and substrate specificity and similar kinetic parameters with A-M1 from Plasmodium falciparum (PfA-M1), a validated target in the treatment of malaria. EtAPN1 is synthesized as a 120-kDa precursor and cleaved into 96-, 68-, and 38-kDa forms during sporulation. Further, immunolocalization assays revealed that, similar to PfA-M1, EtAPN1 is present during the intracellular life cycle stages in both the parasite cytoplasm and the parasite nucleus. The present results support the hypothesis of a conserved role between the two aminopeptidases, and we suggest that EtAPN1 might be a valuable target for anticoccidiosis drugs.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

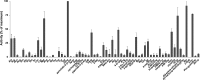
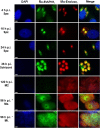
 ) was assayed by treatment of infected MDBK cells with bestatin. For determination of invasion, cells were fixed at 4 h p.i. and the percent invasion was calculated. For determination of development, supernatants of the cell culture were collected at 96 h p.i. and the number of first-generation merozoites was estimated and compared with that for a control without inhibitor. Values are reported as the mean ± SD of three independent experiments. Intergroup differences (P values) between the control and treated conditions were evaluated by Student's paired t test, and significance is indicated (***, P < 0.001). (B) MDBK cells infected with Wis YFP+ sporozoites were fixed at 72 h p.i. and mounted in Vectashield mounting medium. Cell nuclei were labeled with DAPI (blue). YFP+ parasites appear green. Images are representative of those from three individual experiments. Bar, 2 μm.
) was assayed by treatment of infected MDBK cells with bestatin. For determination of invasion, cells were fixed at 4 h p.i. and the percent invasion was calculated. For determination of development, supernatants of the cell culture were collected at 96 h p.i. and the number of first-generation merozoites was estimated and compared with that for a control without inhibitor. Values are reported as the mean ± SD of three independent experiments. Intergroup differences (P values) between the control and treated conditions were evaluated by Student's paired t test, and significance is indicated (***, P < 0.001). (B) MDBK cells infected with Wis YFP+ sporozoites were fixed at 72 h p.i. and mounted in Vectashield mounting medium. Cell nuclei were labeled with DAPI (blue). YFP+ parasites appear green. Images are representative of those from three individual experiments. Bar, 2 μm.
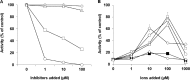
 ), and amastatin (■) against unsporulated oocyst lysate was assayed. The lysate was preincubated with the inhibitor before addition of the H-Ala-AMC. Values are reported as the mean ± SD of three independent experiments. Intergroup differences (P values) between the control and treated conditions were evaluated by Student's paired t test, and significance is indicated (§, P < 0.001 for 1,10-ortho-phenantroline; #, P < 0.05 for bestatin; *, P < 0.001 for amastatin).
), and amastatin (■) against unsporulated oocyst lysate was assayed. The lysate was preincubated with the inhibitor before addition of the H-Ala-AMC. Values are reported as the mean ± SD of three independent experiments. Intergroup differences (P values) between the control and treated conditions were evaluated by Student's paired t test, and significance is indicated (§, P < 0.001 for 1,10-ortho-phenantroline; #, P < 0.05 for bestatin; *, P < 0.001 for amastatin).


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

