A knowledge-driven approach to extract disease-related biomarkers from the literature
- PMID: 24839601
- PMCID: PMC4009255
- DOI: 10.1155/2014/253128
A knowledge-driven approach to extract disease-related biomarkers from the literature
Abstract
The biomedical literature represents a rich source of biomarker information. However, both the size of literature databases and their lack of standardization hamper the automatic exploitation of the information contained in these resources. Text mining approaches have proven to be useful for the exploitation of information contained in the scientific publications. Here, we show that a knowledge-driven text mining approach can exploit a large literature database to extract a dataset of biomarkers related to diseases covering all therapeutic areas. Our methodology takes advantage of the annotation of MEDLINE publications pertaining to biomarkers with MeSH terms, narrowing the search to specific publications and, therefore, minimizing the false positive ratio. It is based on a dictionary-based named entity recognition system and a relation extraction module. The application of this methodology resulted in the identification of 131,012 disease-biomarker associations between 2,803 genes and 2,751 diseases, and represents a valuable knowledge base for those interested in disease-related biomarkers. Additionally, we present a bibliometric analysis of the journals reporting biomarker related information during the last 40 years.
Figures

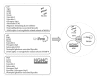
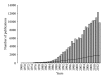
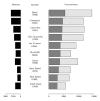
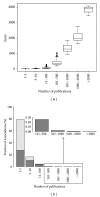
References
-
- Atkinson AJ, Colburn WA, deGruttola VG, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical Pharmacology & Therapeutics. 2001;69(3):89–95. - PubMed
-
- Guidance for Industry-E15 Definitions for Genomic Biomarkers, Pharmacogenomics, Pharmacogenetics, Genomic Data and Sample Coding Categories, http://www.fda.gov/downloads/RegulatoryInformation/Guidances/ucm129296.pdf.
-
- Anderson DC, Kodukula K. Biomarkers in pharmacology and drug discovery. Biochemical Pharmacology. 2014;87(1):172–188. - PubMed
-
- Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nature Reviews Drug Discovery. 2003;2(7):566–580. - PubMed
-
- Dancey JE, Dobbin KK, Groshen S, et al. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clinical Cancer Research. 2010;16(6):1745–1755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

