Mg-chelatase I subunit 1 and Mg-protoporphyrin IX methyltransferase affect the stomatal aperture in Arabidopsis thaliana
- PMID: 24840863
- PMCID: PMC4683165
- DOI: 10.1007/s10265-014-0636-0
Mg-chelatase I subunit 1 and Mg-protoporphyrin IX methyltransferase affect the stomatal aperture in Arabidopsis thaliana
Abstract
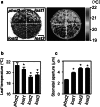
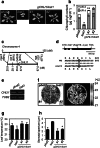
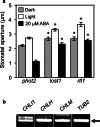
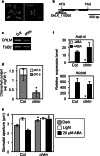
To elucidate the molecular mechanisms of stomatal opening and closure, we performed a genetic screen using infrared thermography to isolate stomatal aperture mutants. We identified a mutant designated low temperature with open-stomata 1 (lost1), which exhibited reduced leaf temperature, wider stomatal aperture, and a pale green phenotype. Map-based analysis of the LOST1 locus revealed that the lost1 mutant resulted from a missense mutation in the Mg-chelatase I subunit 1 (CHLI1) gene, which encodes a subunit of the Mg-chelatase complex involved in chlorophyll synthesis. Transformation of the wild-type CHLI1 gene into lost1 complemented all lost1 phenotypes. Stomata in lost1 exhibited a partial ABA-insensitive phenotype similar to that of rtl1, a Mg-chelatase H subunit missense mutant. The Mg-protoporphyrin IX methyltransferase (CHLM) gene encodes a subsequent enzyme in the chlorophyll synthesis pathway. We examined stomatal movement in a CHLM knockdown mutant, chlm, and found that it also exhibited an ABA-insensitive phenotype. However, lost1 and chlm seedlings all showed normal expression of ABA-induced genes, such as RAB18 and RD29B, in response to ABA. These results suggest that the chlorophyll synthesis enzymes, Mg-chelatase complex and CHLM, specifically affect ABA signaling in the control of stomatal aperture and have no effect on ABA-induced gene expression.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

