Proteolytic activation of the human epithelial sodium channel by trypsin IV and trypsin I involves distinct cleavage sites
- PMID: 24841206
- PMCID: PMC4081944
- DOI: 10.1074/jbc.M113.538470
Proteolytic activation of the human epithelial sodium channel by trypsin IV and trypsin I involves distinct cleavage sites
Abstract
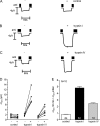
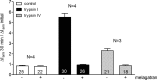
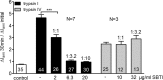
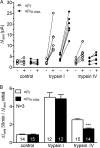
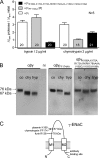
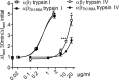
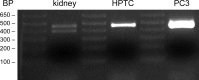
Proteolytic activation is a unique feature of the epithelial sodium channel (ENaC). However, the underlying molecular mechanisms and the physiologically relevant proteases remain to be identified. The serine protease trypsin I can activate ENaC in vitro but is unlikely to be the physiologically relevant activating protease in ENaC-expressing tissues in vivo. Herein, we investigated whether human trypsin IV, a form of trypsin that is co-expressed in several extrapancreatic epithelial cells with ENaC, can activate human ENaC. In Xenopus laevis oocytes, we monitored proteolytic activation of ENaC currents and the appearance of γENaC cleavage products at the cell surface. We demonstrated that trypsin IV and trypsin I can stimulate ENaC heterologously expressed in oocytes. ENaC cleavage and activation by trypsin IV but not by trypsin I required a critical cleavage site (Lys-189) in the extracellular domain of the γ-subunit. In contrast, channel activation by trypsin I was prevented by mutating three putative cleavage sites (Lys-168, Lys-170, and Arg-172) in addition to mutating previously described prostasin (RKRK(178)), plasmin (Lys-189), and neutrophil elastase (Val-182 and Val-193) sites. Moreover, we found that trypsin IV is expressed in human renal epithelial cells and can increase ENaC-mediated sodium transport in cultured human airway epithelial cells. Thus, trypsin IV may regulate ENaC function in epithelial tissues. Our results show, for the first time, that trypsin IV can stimulate ENaC and that trypsin IV and trypsin I activate ENaC by cleavage at distinct sites. The presence of distinct cleavage sites may be important for ENaC regulation by tissue-specific proteases.
Keywords: Epithelial Sodium Channel (ENaC); Ion Channel; Oocyte; Protease; Proteolytic Channel Activation; Serine Protease; Trypsin; Trypsin IV; Two-electrode Voltage Clamp.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Garty H., Palmer L. G. (1997) Epithelial sodium channels: function, structure, and regulation. Physiol. Rev. 77, 359–396 - PubMed
-
- Kellenberger S., Schild L. (2002) Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol. Rev. 82, 735–767 - PubMed
-
- Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 - PubMed
-
- Stockand J. D., Staruschenko A., Pochynyuk O., Booth R. E., Silverthorn D. U. (2008) Insight toward epithelial Na+ channel mechanism revealed by the acid-sensing ion channel 1 structure. IUBMB Life 60, 620–628 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

