In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment
- PMID: 24841267
- PMCID: PMC4136067
- DOI: 10.1128/AAC.02555-14
In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment
Abstract
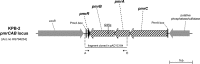
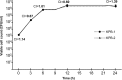
Colistin is a key drug for the treatment of infections caused by extensively drug-resistant strains of Enterobacteriaceae producing carbapenemases. However, the emergence of colistin resistance is being increasingly reported, especially among Klebsiella pneumoniae strains producing KPC-type carbapenemases (KPC-KP). In this work, we investigated colistin-susceptible (KPB-1) and colistin-resistant (KPB-2) sequential isolates obtained from a patient with a KPC-KP infection before and after low-dosage colistin treatment, respectively. By using a next-generation sequencing approach and comparative genomic analysis of the two isolates, we detected in KPB-2 a nonsynonymous nucleotide substitution in the gene encoding the PmrB sensor kinase, resulting in a leucine-to-arginine substitution at amino acid position 82. Compared with KPB-1, KPB-2 exhibited upregulated transcription of pmrA and of pmrK, which is part of the pmrHFIJKLM operon responsible for modification of the colistin lipopolysaccharide target. Complementation with wild-type pmrB in KPB-2 restored colistin susceptibility and reduced the transcription of pmrA and pmrK to basal levels, while expression of PmrB(L82R) in KPB-1 did not alter colistin susceptibility or upregulate pmrA and pmrK expression, confirming the dominance of wild-type PmrB versus the PmrB(L82R) mutant. The present results indicated that PmrB mutations mediating colistin resistance may be selected during low-dosage colistin treatment. The colistin-resistant phenotype of KPB-2 was stable for up to 50 generations in the absence of selective pressure and was not associated with a significant fitness cost in a competition experiment.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Canton R, Akova M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P, European Network on Carbapenemases 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431. 10.1111/j.1469-0691.2012.03821.x - DOI - PubMed
-
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 - DOI - PMC - PubMed
-
- Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, AMCLI-CRE Survey Participants. Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill. 18:pii=20489 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20489 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

