Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses
- PMID: 24841268
- PMCID: PMC4136041
- DOI: 10.1128/AAC.02994-14
Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses
Abstract
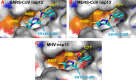
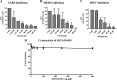
We have previously shown that SSYA10-001 blocks severe acute respiratory syndrome coronavirus (SARS-CoV) replication by inhibiting SARS-CoV helicase (nsp13). Here, we show that SSYA10-001 also inhibits replication of two other coronaviruses, mouse hepatitis virus (MHV) and Middle Eastern respiratory syndrome coronavirus (MERS-CoV). A putative binding pocket for SSYA10-001 was identified and shown to be similar in SARS-CoV, MERS-CoV, and MHV helicases. These studies show that it is possible to target multiple coronaviruses through broad-spectrum inhibitors.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M, Chan AK, Skowronski DM, Salit I, Simor AE, Slutsky AS, Doyle PW, Krajden M, Petric M, Brunham RC, McGeer AJ. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995–2005. 10.1056/NEJMoa030634 - DOI - PubMed
-
- Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. 10.1038/nature12005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM103368/GM/NIGMS NIH HHS/United States
- GM103368/GM/NIGMS NIH HHS/United States
- R21 AI079801/AI/NIAID NIH HHS/United States
- AI076119/AI/NIAID NIH HHS/United States
- R01 AI100890/AI/NIAID NIH HHS/United States
- AI099284/AI/NIAID NIH HHS/United States
- AI112417/AI/NIAID NIH HHS/United States
- R01 AI095569/AI/NIAID NIH HHS/United States
- AI100890/AI/NIAID NIH HHS/United States
- R33 AI079801/AI/NIAID NIH HHS/United States
- AI079801/AI/NIAID NIH HHS/United States
- R37 AI076119/AI/NIAID NIH HHS/United States
- R01 AI099284/AI/NIAID NIH HHS/United States
- R21 AI112417/AI/NIAID NIH HHS/United States
- R01 AI076119/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

