Sandwich antibody arrays using recombinant antibody-binding protein L
- PMID: 24841983
- PMCID: PMC4059220
- DOI: 10.1021/la500822w
Sandwich antibody arrays using recombinant antibody-binding protein L
Abstract
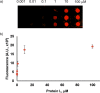
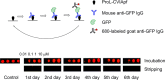
Antibody arrays are a useful for detecting antigens and other antibodies. This technique typically requires a uniform and well-defined orientation of antibodies attached to a surface for optimal performance. A uniform orientation can be achieved by modification of antibodies to include a single site for attachment. Thus, uniformly oriented antibody arrays require a bioengineered modification for the antibodies directly immobilization on the solid surface. In this study, we describe a "sandwich-type" antibody array where unmodified antibodies are oriented through binding with regioselectively immobilized recombinant antibody-binding protein L. Recombinant proL-CVIA bearing C-terminal CVIA motif is post-translationally modified with an alkyne group by protein farnesyltransferase (PFTase) at the cysteine residue in the CVIA sequence to give proL-CVIApf, which is covalently attached to an azido-modified glass slide by a Huisgen [3 + 2] cycloaddition reaction. Slides bearing antibodies bound to slides coated with regioselectively immobilized proL-CVIApf gave stronger fluorescence outputs and those where the antibody-binding protein was immobilized in random orientations on an epoxy-modified slide. Properly selected capture and detection antibodies did not cross-react with immobilized proL-CVIApf in sandwich arrays, and the proL-CVIApf slides can be used for multiple cycles of detected over a period of several months.
Figures






Similar articles
-
Regioselective covalent immobilization of recombinant antibody-binding proteins A, G, and L for construction of antibody arrays.J Am Chem Soc. 2013 Jun 19;135(24):8973-80. doi: 10.1021/ja402447g. Epub 2013 Jun 10. J Am Chem Soc. 2013. PMID: 23746333 Free PMC article.
-
Regioselective covalent immobilization of catalytically active glutathione S-transferase on glass slides.Bioconjug Chem. 2013 Apr 17;24(4):571-7. doi: 10.1021/bc300462j. Epub 2013 Mar 26. Bioconjug Chem. 2013. PMID: 23458569 Free PMC article.
-
Evaluation of homo- and hetero-functionally activated glass surfaces for optimized antibody arrays.Anal Biochem. 2014 Apr 1;450:37-45. doi: 10.1016/j.ab.2014.01.002. Epub 2014 Jan 15. Anal Biochem. 2014. PMID: 24440232
-
Site-directed antibody immobilization techniques for immunosensors.Biosens Bioelectron. 2013 Dec 15;50:460-71. doi: 10.1016/j.bios.2013.06.060. Epub 2013 Jul 5. Biosens Bioelectron. 2013. PMID: 23911661 Review.
-
Orientation and characterization of immobilized antibodies for improved immunoassays (Review).Biointerphases. 2017 Mar 16;12(2):02D301. doi: 10.1116/1.4978435. Biointerphases. 2017. PMID: 28301944 Review.
Cited by
-
Characterization of FcγRIa (CD64) as a Ligand Molecule for Site-Specific IgG1 Capture: A Side-By-Side Comparison with Protein A.Langmuir. 2022 Dec 6;38(48):14623-14634. doi: 10.1021/acs.langmuir.2c02022. Epub 2022 Nov 23. Langmuir. 2022. PMID: 36416530 Free PMC article.
-
Recent progress in enzymatic protein labelling techniques and their applications.Chem Soc Rev. 2018 Dec 21;47(24):9106-9136. doi: 10.1039/c8cs00537k. Epub 2018 Sep 27. Chem Soc Rev. 2018. PMID: 30259933 Free PMC article. Review.
-
A Not-So-Ancient Grease History: Click Chemistry and Protein Lipid Modifications.Chem Rev. 2021 Jun 23;121(12):7178-7248. doi: 10.1021/acs.chemrev.0c01108. Epub 2021 Apr 6. Chem Rev. 2021. PMID: 33821625 Free PMC article. Review.
-
Biofunctionalization of Multiplexed Silicon Photonic Biosensors.Biosensors (Basel). 2022 Dec 29;13(1):53. doi: 10.3390/bios13010053. Biosensors (Basel). 2022. PMID: 36671887 Free PMC article. Review.
-
Protein prenylation: enzymes, therapeutics, and biotechnology applications.ACS Chem Biol. 2015 Jan 16;10(1):51-62. doi: 10.1021/cb500791f. Epub 2014 Dec 8. ACS Chem Biol. 2015. PMID: 25402849 Free PMC article. Review.
References
-
- Rusmini F.; Zhong Z.; Feijen J. Protein immobilization strategies for protein biochips. Biomacromolecules 2007, 8, 1775–1789. - PubMed
-
- Zhang C.-J.; Tan C. Y. J.; Ge J.; Na Z.; Chen G. Y. J.; Uttamchandani M.; Sun H.; Yao S. Q. Preparation of small-molecule microarrays by trans-cyclooctene tetrazine ligation and their application in the high-throughput screening of protein-protein interaction inhibitors of bromodomains. Angew. Chem., Int. Ed. 2013, 52, 14060–14064. - PubMed
-
- Liu Y.; He J.; Yang K.-L. DNA microarrays on ultraviolet-modified surfaces for speciation of bacteria. Anal. Biochem. 2014, 447, 156–161. - PubMed
-
- Blackburn J. M.; Shoko A.; Beeton-Kempen N. Miniaturized, microarray-based assays for chemical proteomic studies of protein function. Methods Mol. Biol. 2012, 800, 133–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

