SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint
- PMID: 24843002
- PMCID: PMC4038841
- DOI: 10.7554/eLife.02482
SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint
Abstract
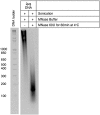
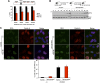
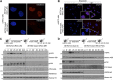
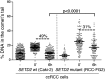
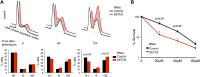
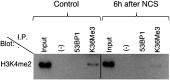
Histone modifications establish the chromatin states that coordinate the DNA damage response. In this study, we show that SETD2, the enzyme that trimethylates histone H3 lysine 36 (H3K36me3), is required for ATM activation upon DNA double-strand breaks (DSBs). Moreover, we find that SETD2 is necessary for homologous recombination repair of DSBs by promoting the formation of RAD51 presynaptic filaments. In agreement, SETD2-mutant clear cell renal cell carcinoma (ccRCC) cells displayed impaired DNA damage signaling. However, despite the persistence of DNA lesions, SETD2-deficient cells failed to activate p53, a master guardian of the genome rarely mutated in ccRCC and showed decreased cell survival after DNA damage. We propose that this novel SETD2-dependent role provides a chromatin bookmarking instrument that facilitates signaling and repair of DSBs. In ccRCC, loss of SETD2 may afford an alternative mechanism for the inactivation of the p53-mediated checkpoint without the need for additional genetic mutations in TP53.DOI: http://dx.doi.org/10.7554/eLife.02482.001.
Keywords: DNA damage response; SETD2; cancer; p53-mediated checkpoint.
Copyright © 2014, Carvalho et al.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






References
-
- Bohm L, Briand G, Sautiere P, Crane-Robinson C. 1981. Proteolytic digestion studies of chromatin core-histone structure. Identification of the limit peptides of histones H3 and H4. European Journal of Biochemistry/FEBS 119:67–74 - PubMed
-
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Structural & Molecular Biology 17:688–695. doi: 10.1038/nsmb.1831 - DOI - PMC - PubMed
-
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. doi: 10.1016/j.cell.2010.03.012 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

