Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci
- PMID: 24843030
- PMCID: PMC4017647
- DOI: 10.7554/eLife.02557
Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci
Abstract
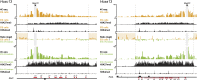
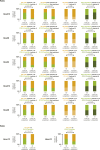
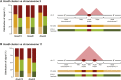
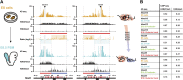
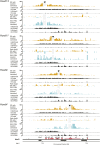
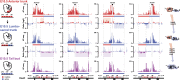
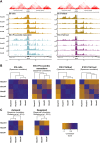
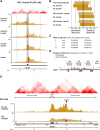
Hox genes are essential regulators of embryonic development. Their step-wise transcriptional activation follows their genomic topology and the various states of activation are subsequently memorized into domains of progressively overlapping gene products. We have analyzed the 3D chromatin organization of Hox clusters during their early activation in vivo, using high-resolution circular chromosome conformation capture. Initially, Hox clusters are organized as single chromatin compartments containing all genes and bivalent chromatin marks. Transcriptional activation is associated with a dynamic bi-modal 3D organization, whereby the genes switch autonomously from an inactive to an active compartment. These local 3D dynamics occur within a framework of constitutive interactions within the surrounding Topological Associated Domains, indicating that this regulation process is mostly cluster intrinsic. The step-wise progression in time is fixed at various body levels and thus can account for the chromatin architectures previously described at a later stage for different anterior to posterior levels.DOI: http://dx.doi.org/10.7554/eLife.02557.001.
Keywords: Hox gene regulation; chromatin domains; gene regulation; topological domains.
Copyright © 2014, Noordermeer et al.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










References
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. doi: 10.1016/j.cell.2006.02.041 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

