Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing
- PMID: 24843150
- PMCID: PMC4060699
- DOI: 10.1073/pnas.1406103111
Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing
Abstract
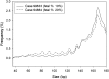
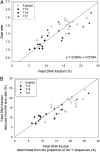
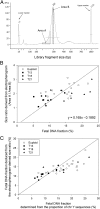
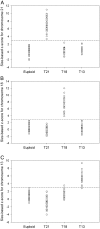
Noninvasive prenatal testing using fetal DNA in maternal plasma is an actively researched area. The current generation of tests using massively parallel sequencing is based on counting plasma DNA sequences originating from different genomic regions. In this study, we explored a different approach that is based on the use of DNA fragment size as a diagnostic parameter. This approach is dependent on the fact that circulating fetal DNA molecules are generally shorter than the corresponding maternal DNA molecules. First, we performed plasma DNA size analysis using paired-end massively parallel sequencing and microchip-based capillary electrophoresis. We demonstrated that the fetal DNA fraction in maternal plasma could be deduced from the overall size distribution of maternal plasma DNA. The fetal DNA fraction is a critical parameter affecting the accuracy of noninvasive prenatal testing using maternal plasma DNA. Second, we showed that fetal chromosomal aneuploidy could be detected by observing an aberrant proportion of short fragments from an aneuploid chromosome in the paired-end sequencing data. Using this approach, we detected fetal trisomy 21 and trisomy 18 with 100% sensitivity (T21: 36/36; T18: 27/27) and 100% specificity (non-T21: 88/88; non-T18: 97/97). For trisomy 13, the sensitivity and specificity were 95.2% (20/21) and 99% (102/103), respectively. For monosomy X, the sensitivity and specificity were both 100% (10/10 and 8/8). Thus, this study establishes the principle of size-based molecular diagnostics using plasma DNA. This approach has potential applications beyond noninvasive prenatal testing to areas such as oncology and transplantation monitoring.
Keywords: Down syndrome; Turner syndrome; fetal aneuploidy; next-generation sequencing; size profiling.
Conflict of interest statement
Conflict of interest statement: R.W.K.C. and Y.M.D.L. have support from Sequenom, Inc. for the submitted work. R.W.K.C. and Y.M.D.L. are consultants to, and hold equities in, Sequenom, Inc. K.C.A.C. holds equities in Sequenom, Inc. The technology reported in this paper is covered by US Patent 8,620,593 and US Patent Application 2013/0237431.
Figures
Similar articles
-
Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center.Ultrasound Obstet Gynecol. 2014 Mar;43(3):254-64. doi: 10.1002/uog.13277. Epub 2014 Feb 10. Ultrasound Obstet Gynecol. 2014. PMID: 24339153 Review.
-
Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing.Am J Obstet Gynecol. 2014 Nov;211(5):527.e1-527.e17. doi: 10.1016/j.ajog.2014.08.006. Epub 2014 Aug 8. Am J Obstet Gynecol. 2014. PMID: 25111587
-
DNA sequencing versus standard prenatal aneuploidy screening.N Engl J Med. 2014 Feb 27;370(9):799-808. doi: 10.1056/NEJMoa1311037. N Engl J Med. 2014. PMID: 24571752
-
Performance of the neoBona test: a new paired-end massively parallel shotgun sequencing approach for cell-free DNA-based aneuploidy screening.Ultrasound Obstet Gynecol. 2017 Apr;49(4):460-464. doi: 10.1002/uog.17386. Epub 2017 Feb 28. Ultrasound Obstet Gynecol. 2017. PMID: 27981672 Free PMC article.
-
Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis.Ultrasound Obstet Gynecol. 2015 Mar;45(3):249-66. doi: 10.1002/uog.14791. Epub 2015 Feb 1. Ultrasound Obstet Gynecol. 2015. Update in: Ultrasound Obstet Gynecol. 2017 Sep;50(3):302-314. doi: 10.1002/uog.17484. PMID: 25639627 Updated. Review.
Cited by
-
Detection and characterization of jagged ends of double-stranded DNA in plasma.Genome Res. 2020 Aug;30(8):1144-1153. doi: 10.1101/gr.261396.120. Epub 2020 Aug 14. Genome Res. 2020. PMID: 32801148 Free PMC article.
-
Sequencing of short cfDNA fragments in NIPT improves fetal fraction with higher maternal BMI and early gestational age.Am J Transl Res. 2019 Jul 15;11(7):4450-4459. eCollection 2019. Am J Transl Res. 2019. PMID: 31396348 Free PMC article.
-
Cell-Free DNA Fragmentomics in Liquid Biopsy.Diagnostics (Basel). 2022 Apr 13;12(4):978. doi: 10.3390/diagnostics12040978. Diagnostics (Basel). 2022. PMID: 35454026 Free PMC article. Review.
-
FF-QuantSC: accurate quantification of fetal fraction by a neural network model.Mol Genet Genomic Med. 2020 Jun;8(6):e1232. doi: 10.1002/mgg3.1232. Epub 2020 Apr 13. Mol Genet Genomic Med. 2020. PMID: 32281746 Free PMC article.
-
Emerging frontiers of cell-free DNA fragmentomics.Extracell Vesicles Circ Nucl Acids. 2022 Dec 21;3(4):380-392. doi: 10.20517/evcna.2022.34. eCollection 2022. Extracell Vesicles Circ Nucl Acids. 2022. PMID: 39697357 Free PMC article. Review.
References
-
- Lo YMD, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. - PubMed
-
- Chan KCA, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50(1):88–92. - PubMed
-
- Chan KCA, Yeung SW, Lui WB, Rainer TH, Lo YMD. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin Chem. 2005;51(4):781–784. - PubMed
-
- Li Y, et al. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem. 2004;50(6):1002–1011. - PubMed
-
- Li Y, et al. Detection of paternally inherited fetal point mutations for beta-thalassemia using size-fractionated cell-free DNA in maternal plasma. JAMA. 2005;293(7):843–849. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

