Removing T-cell epitopes with computational protein design
- PMID: 24843166
- PMCID: PMC4060723
- DOI: 10.1073/pnas.1321126111
Removing T-cell epitopes with computational protein design
Abstract
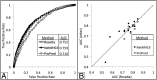
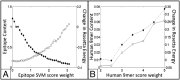
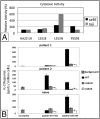
Immune responses can make protein therapeutics ineffective or even dangerous. We describe a general computational protein design method for reducing immunogenicity by eliminating known and predicted T-cell epitopes and maximizing the content of human peptide sequences without disrupting protein structure and function. We show that the method recapitulates previous experimental results on immunogenicity reduction, and we use it to disrupt T-cell epitopes in GFP and Pseudomonas exotoxin A without disrupting function.
Keywords: Rosetta; biotherapeutics; deimmunization; immunotoxin; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zubler RH. Naive and Memory B Cells in T-Cell-Dependent and T-Independent Responses. Springer Seminars in Immunopathology. Berlin: Springer; 2001. pp. 405–419. - PubMed
-
- Goldenberg MM. Trastuzumab, a recombinant DNA-derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin Ther. 1999;21(2):309–318. - PubMed
-
- Harding FA, et al. A beta-lactamase with reduced immunogenicity for the targeted delivery of chemotherapeutics using antibody-directed enzyme prodrug therapy. Mol Cancer Ther. 2005;4(11):1791–1800. - PubMed
-
- Tangri S, et al. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174(6):3187–3196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

