Insulin gene mutations and diabetes
- PMID: 24843467
- PMCID: PMC4015537
- DOI: 10.1111/j.2040-1124.2011.00100.x
Insulin gene mutations and diabetes
Abstract
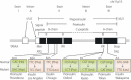
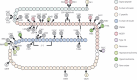
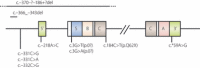
Some mutations of the insulin gene cause hyperinsulinemia or hyperproinsulinemia. Replacement of biologically important amino acid leads to defective receptor binding, longer half-life and hyperinsulinemia. Three mutant insulins have been identified: (i) insulin Chicago (F49L or PheB25Leu); (ii) insulin Los Angeles (F48S or PheB24Ser); (iii) and insulin Wakayama (V92L or ValA3Leu). Replacement of amino acid is necessary for proinsulin processing results in hyperproinsulinemia. Four types have been identified: (i) proinsulin Providence (H34D); (ii) proinsulin Tokyo (R89H); (iii) proinsulin Kyoto (R89L); and (iv) proinsulin Oxford (R89P). Three of these are processing site mutations. The mutation of proinsulin Providence, in contrast, is thought to cause sorting abnormality. Compared with normal proinsulin, a significant amount of proinsulin Providence enters the constitutive pathway where processing does not occur. These insulin gene mutations with hyper(pro)insulinemia were very rare, showed only mild diabetes or glucose intolerance, and hyper(pro)insulinemia was the key for their diagnosis. However, this situation changed dramatically after the identification of insulin gene mutations as a cause of neonatal diabetes. This class of insulin gene mutations does not show hyper(pro)insulinemia. Mutations at the cysteine residue or creating a new cysteine will disturb the correct disulfide bonding and proper conformation, and finally will lead to misfolded proinsulin accumulation, endoplasmic reticulum stress and apoptosis of pancreatic β-cells. Maturity-onset diabetes of the young (MODY) or an autoantibody-negative type 1-like phenotype has also been reported. Very recently, recessive mutations with reduced insulin biosynthesis have been reported. The importance of insulin gene mutation in the pathogenesis of diabetes will increase a great deal and give us a new understanding of β-cell biology and diabetes. (J Diabetes Invest, doi: 10.1111/j.2040-1124.2011.00100.x, 2011).
Keywords: Endoplasmic reticulum stress; Insulin gene mutation; Neonatal diabetes.
Figures
Similar articles
-
Biological behaviors of mutant proinsulin contribute to the phenotypic spectrum of diabetes associated with insulin gene mutations.Mol Cell Endocrinol. 2020 Dec 1;518:111025. doi: 10.1016/j.mce.2020.111025. Epub 2020 Sep 8. Mol Cell Endocrinol. 2020. PMID: 32916194 Free PMC article.
-
Mutant proinsulin proteins associated with neonatal diabetes are retained in the endoplasmic reticulum and not efficiently secreted.Biochem Biophys Res Commun. 2010 Jan 15;391(3):1449-54. doi: 10.1016/j.bbrc.2009.12.090. Epub 2009 Dec 23. Biochem Biophys Res Commun. 2010. PMID: 20034470 Free PMC article.
-
Lessons learned from molecular biology of insulin-gene mutations.Diabetes Care. 1990 Jun;13(6):600-9. doi: 10.2337/diacare.13.6.600. Diabetes Care. 1990. PMID: 2192846 Review.
-
Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on wild-type proinsulin transport.PLoS One. 2010 Oct 11;5(10):e13333. doi: 10.1371/journal.pone.0013333. PLoS One. 2010. PMID: 20948967 Free PMC article.
-
Hyperproinsulinemia in a three-generation Caucasian family due to mutant proinsulin (Arg65-His) not associated with imparied glucose tolerance: the contribution of mutant proinsulin to insulin bioactivity.J Clin Endocrinol Metab. 1996 Apr;81(4):1634-40. doi: 10.1210/jcem.81.4.8636380. J Clin Endocrinol Metab. 1996. PMID: 8636380 Review.
Cited by
-
Type 1 Diabetes Mellitus: A Review on Advances and Challenges in Creating Insulin Producing Devices.Micromachines (Basel). 2023 Jan 6;14(1):151. doi: 10.3390/mi14010151. Micromachines (Basel). 2023. PMID: 36677212 Free PMC article. Review.
-
Exploring proinsulin proteostasis: insights into beta cell health and diabetes.Front Mol Biosci. 2025 Mar 5;12:1554717. doi: 10.3389/fmolb.2025.1554717. eCollection 2025. Front Mol Biosci. 2025. PMID: 40109403 Free PMC article. Review.
-
Genetic Etiology of Neonatal Diabetes Mellitus in Vietnamese Infants and Characteristics of Those With INS Gene Mutations.Front Endocrinol (Lausanne). 2022 Apr 19;13:866573. doi: 10.3389/fendo.2022.866573. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35518939 Free PMC article.
-
Structural and functional study of the GlnB22-insulin mutant responsible for maturity-onset diabetes of the young.PLoS One. 2014 Nov 25;9(11):e112883. doi: 10.1371/journal.pone.0112883. eCollection 2014. PLoS One. 2014. PMID: 25423173 Free PMC article.
-
Maturity-Onset Diabetes of the Young: Mutations, Physiological Consequences, and Treatment Options.J Pers Med. 2022 Oct 25;12(11):1762. doi: 10.3390/jpm12111762. J Pers Med. 2022. PMID: 36573710 Free PMC article. Review.
References
-
- Tager H, Given B, Baldwin B, et al. A structurally abnormal insulin causing human diabetes. Nature 1979; 281: 122–125 - PubMed
-
- Given BD, Mako ME, Tager HS, et al. Diabetes due to secretion of an abnormal insulin. N Engl J Med 1980; 302: 129–135 - PubMed
-
- Shoelson S, Haneda M, Blix P, et al. Three mutant insulin in man. Nature 1983; 302: 540–543 - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

