An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK
- PMID: 24844597
- PMCID: PMC4028294
- DOI: 10.1371/journal.pone.0098103
An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK
Abstract
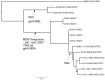
Following a large outbreak of foodborne gastrointestinal (GI) disease, a multiplex PCR approach was used retrospectively to investigate faecal specimens from 88 of the 413 reported cases. Gene targets from a range of bacterial GI pathogens were detected, including Salmonella species, Shigella species and Shiga toxin-producing Escherichia coli, with the majority (75%) of faecal specimens being PCR positive for aggR associated with the Enteroaggregative E. coli (EAEC) group. The 20 isolates of EAEC recovered from the outbreak specimens exhibited a range of serotypes, the most frequent being O104:H4 and O131:H27. None of the EAEC isolates had the Shiga toxin (stx) genes. Multilocus sequence typing and single nucleotide polymorphism analysis of the core genome confirmed the diverse phylogeny of the strains. The analysis also revealed a close phylogenetic relationship between the EAEC O104:H4 strains in this outbreak and the strain of E. coli O104:H4 associated with a large outbreak of haemolytic ureamic syndrome in Germany in 2011. Further analysis of the EAEC plasmids, encoding the key enteroaggregative virulence genes, showed diversity with respect to FIB/FII type, gene content and genomic architecture. Known EAEC virulence genes, such as aggR, aat and aap, were present in all but one of the strains. A variety of fimbrial genes were observed, including genes encoding all five known fimbrial types, AAF/1 to AAF/V. The AAI operon was present in its entirety in 15 of the EAEC strains, absent in three and present, but incomplete, in two isolates. EAEC is known to be a diverse pathotype and this study demonstrates that a high level of diversity in strains recovered from cases associated with a single outbreak. Although the EAEC in this study did not carry the stx genes, this outbreak provides further evidence of the pathogenic potential of the EAEC O104:H4 serotype.
Conflict of interest statement
Figures


Similar articles
-
Escherichia coli O104:H4 Pathogenesis: an Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence.Microbiol Spectr. 2014 Dec;2(6). doi: 10.1128/microbiolspec.EHEC-0008-2013. Microbiol Spectr. 2014. PMID: 26104460 Review.
-
Comparative virulence characterization of the Shiga toxin phage-cured Escherichia coli O104:H4 and enteroaggregative Escherichia coli.Int J Med Microbiol. 2018 Oct;308(7):912-920. doi: 10.1016/j.ijmm.2018.06.006. Epub 2018 Jun 19. Int J Med Microbiol. 2018. PMID: 29941383
-
Characteristics of the Shiga-toxin-producing enteroaggregative Escherichia coli O104:H4 German outbreak strain and of STEC strains isolated in Spain.Int Microbiol. 2011 Sep;14(3):121-41. doi: 10.2436/20.1501.01.142. Int Microbiol. 2011. PMID: 22101411 Review.
-
Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study.Lancet Infect Dis. 2011 Sep;11(9):671-6. doi: 10.1016/S1473-3099(11)70165-7. Epub 2011 Jun 22. Lancet Infect Dis. 2011. PMID: 21703928
-
Colonization of Enteroaggregative Escherichia coli and Shiga toxin-producing Escherichia coli in chickens and humans in southern Vietnam.BMC Microbiol. 2016 Sep 9;16(1):208. doi: 10.1186/s12866-016-0827-z. BMC Microbiol. 2016. PMID: 27612880 Free PMC article.
Cited by
-
Shiga toxin 2A-encoding bacteriophages in enteroaggregative Escherichia coli O104:H4 strains.Emerg Infect Dis. 2014 Sep;20(9):1567-8. doi: 10.3201/eid2009.131373. Emerg Infect Dis. 2014. PMID: 25152038 Free PMC article. No abstract available.
-
Multipathogen Outbreak of Bacillus cereus and Clostridium perfringens Among Hospital Workers in Alaska, August 2021.Public Health Rep. 2024 Mar-Apr;139(2):195-200. doi: 10.1177/00333549231170220. Epub 2023 May 13. Public Health Rep. 2024. PMID: 37178053 Free PMC article.
-
Large outbreak of multiple gastrointestinal pathogens associated with fresh curry leaves in North East England, 2013.Epidemiol Infect. 2018 Nov;146(15):1940-1947. doi: 10.1017/S095026881800225X. Epub 2018 Aug 15. Epidemiol Infect. 2018. PMID: 30109832 Free PMC article.
-
To kill or to be killed: pangenome analysis of Escherichia coli strains reveals a tailocin specific for pandemic ST131.BMC Biol. 2022 Jun 16;20(1):146. doi: 10.1186/s12915-022-01347-7. BMC Biol. 2022. PMID: 35710371 Free PMC article.
-
Enteroaggregative Escherichia coli in Daycare-A 1-Year Dynamic Cohort Study.Front Cell Infect Microbiol. 2016 Jul 13;6:75. doi: 10.3389/fcimb.2016.00075. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27468409 Free PMC article.
References
-
- Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, et al. (1987) Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J 6: 829–31. - PubMed
-
- Nishi J, Sheikh J, Mizuguchi K, Luisi B, Burland V, et al. (2003) The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J Biol Chem 278: 45680–9. - PubMed
-
- Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP (2006) Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli . Mol Microbiol 61: 1267–82. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

