CLImAT: accurate detection of copy number alteration and loss of heterozygosity in impure and aneuploid tumor samples using whole-genome sequencing data
- PMID: 24845652
- PMCID: PMC4155249
- DOI: 10.1093/bioinformatics/btu346
CLImAT: accurate detection of copy number alteration and loss of heterozygosity in impure and aneuploid tumor samples using whole-genome sequencing data
Abstract
Motivation: Whole-genome sequencing of tumor samples has been demonstrated as an efficient approach for comprehensive analysis of genomic aberrations in cancer genome. Critical issues such as tumor impurity and aneuploidy, GC-content and mappability bias have been reported to complicate identification of copy number alteration and loss of heterozygosity in complex tumor samples. Therefore, efficient computational methods are required to address these issues.
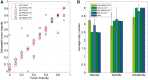
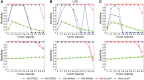
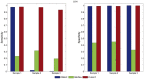
Results: We introduce CLImAT (CNA and LOH Assessment in Impure and Aneuploid Tumors), a bioinformatics tool for identification of genomic aberrations from tumor samples using whole-genome sequencing data. Without requiring a matched normal sample, CLImAT takes integrated analysis of read depth and allelic frequency and provides extensive data processing procedures including GC-content and mappability correction of read depth and quantile normalization of B-allele frequency. CLImAT accurately identifies copy number alteration and loss of heterozygosity even for highly impure tumor samples with aneuploidy. We evaluate CLImAT on both simulated and real DNA sequencing data to demonstrate its ability to infer tumor impurity and ploidy and identify genomic aberrations in complex tumor samples.
Availability and implementation: The CLImAT software package can be freely downloaded at http://bioinformatics.ustc.edu.cn/CLImAT/.
© The Author 2014. Published by Oxford University Press.
Figures

References
-
- Albertson DG, et al. Chromosome aberrations in solid tumors. Nat. Genet. 2003;34:369–376. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

