Microbial community profiling of human saliva using shotgun metagenomic sequencing
- PMID: 24846174
- PMCID: PMC4028220
- DOI: 10.1371/journal.pone.0097699
Microbial community profiling of human saliva using shotgun metagenomic sequencing
Erratum in
- PLoS One. 2014;9(8):e106124
Abstract
Human saliva is clinically informative of both oral and general health. Since next generation shotgun sequencing (NGS) is now widely used to identify and quantify bacteria, we investigated the bacterial flora of saliva microbiomes of two healthy volunteers and five datasets from the Human Microbiome Project, along with a control dataset containing short NGS reads from bacterial species representative of the bacterial flora of human saliva. GENIUS, a system designed to identify and quantify bacterial species using unassembled short NGS reads was used to identify the bacterial species comprising the microbiomes of the saliva samples and datasets. Results, achieved within minutes and at greater than 90% accuracy, showed more than 175 bacterial species comprised the bacterial flora of human saliva, including bacteria known to be commensal human flora but also Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae, and Gamma proteobacteria. Basic Local Alignment Search Tool (BLASTn) analysis in parallel, reported ca. five times more species than those actually comprising the in silico sample. Both GENIUS and BLAST analyses of saliva samples identified major genera comprising the bacterial flora of saliva, but GENIUS provided a more precise description of species composition, identifying to strain in most cases and delivered results at least 10,000 times faster. Therefore, GENIUS offers a facile and accurate system for identification and quantification of bacterial species and/or strains in metagenomic samples.
Conflict of interest statement
Figures

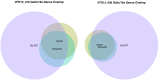



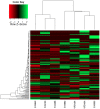
Similar articles
-
Assessing saliva microbiome collection and processing methods.NPJ Biofilms Microbiomes. 2021 Nov 18;7(1):81. doi: 10.1038/s41522-021-00254-z. NPJ Biofilms Microbiomes. 2021. PMID: 34795298 Free PMC article.
-
Core of the saliva microbiome: an analysis of the MG-RAST data.BMC Oral Health. 2021 Jul 16;21(1):351. doi: 10.1186/s12903-021-01719-5. BMC Oral Health. 2021. PMID: 34271900 Free PMC article.
-
Host DNA Depletion in Saliva Samples for Improved Shotgun Metagenomics.Methods Mol Biol. 2021;2327:87-92. doi: 10.1007/978-1-0716-1518-8_6. Methods Mol Biol. 2021. PMID: 34410641
-
An Introduction to Whole-Metagenome Shotgun Sequencing Studies.Methods Mol Biol. 2021;2243:107-122. doi: 10.1007/978-1-0716-1103-6_6. Methods Mol Biol. 2021. PMID: 33606255 Review.
-
Metagenomics methods for the study of plant-associated microbial communities: A review.J Microbiol Methods. 2020 Mar;170:105860. doi: 10.1016/j.mimet.2020.105860. Epub 2020 Feb 4. J Microbiol Methods. 2020. PMID: 32027927 Review.
Cited by
-
A Review on Microbial Species for Forensic Body Fluid Identification in Healthy and Diseased Humans.Curr Microbiol. 2023 Jul 25;80(9):299. doi: 10.1007/s00284-023-03413-x. Curr Microbiol. 2023. PMID: 37491404 Free PMC article. Review.
-
The salivary microbiota in health and disease.J Oral Microbiol. 2020 Feb 4;12(1):1723975. doi: 10.1080/20002297.2020.1723975. eCollection 2020. J Oral Microbiol. 2020. PMID: 32128039 Free PMC article. Review.
-
Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak.BMC Microbiol. 2016 Nov 16;16(1):275. doi: 10.1186/s12866-016-0894-1. BMC Microbiol. 2016. PMID: 27852235 Free PMC article.
-
Advances in Research on Pig Salivary Analytes: A Window to Reveal Pig Health and Physiological Status.Animals (Basel). 2024 Jan 24;14(3):374. doi: 10.3390/ani14030374. Animals (Basel). 2024. PMID: 38338017 Free PMC article. Review.
-
Malaria disrupts the rhesus macaque gut microbiome.Front Cell Infect Microbiol. 2023 Jan 13;12:1058926. doi: 10.3389/fcimb.2022.1058926. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36710962 Free PMC article.
References
-
- Socransky SS, Haffajee AD, Smith GL, Dzink JL (1987) Difficulties encountered in the search for the etiologic agents of destructive periodontal diseases. J Clin Periodontol 14: 588–593. - PubMed
-
- Haffajee AD, Socransky SS (1994) Microbial etiological agents of destructive periodontal diseases. Periodontol 2000 5: 78–111. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

