Integrons: past, present, and future
- PMID: 24847022
- PMCID: PMC4054258
- DOI: 10.1128/MMBR.00056-13
Integrons: past, present, and future
Abstract
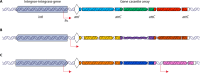
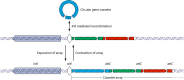
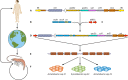
Integrons are versatile gene acquisition systems commonly found in bacterial genomes. They are ancient elements that are a hot spot for genomic complexity, generating phenotypic diversity and shaping adaptive responses. In recent times, they have had a major role in the acquisition, expression, and dissemination of antibiotic resistance genes. Assessing the ongoing threats posed by integrons requires an understanding of their origins and evolutionary history. This review examines the functions and activities of integrons before the antibiotic era. It shows how antibiotic use selected particular integrons from among the environmental pool of these elements, such that integrons carrying resistance genes are now present in the majority of Gram-negative pathogens. Finally, it examines the potential consequences of widespread pollution with the novel integrons that have been assembled via the agency of human antibiotic use and speculates on the potential uses of integrons as platforms for biotechnology.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Partridge SR, Recchia GD, Scaramuzzi C, Collis CM, Stokes H, Hall RM. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855–2864 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

