Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield
- PMID: 24847714
- PMCID: PMC4029903
- DOI: 10.1371/journal.pone.0098264
Transgenic tobacco overexpressing Brassica juncea HMG-CoA synthase 1 shows increased plant growth, pod size and seed yield
Erratum in
- PLoS One. 2014;9(9):e108026
Abstract
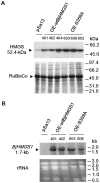
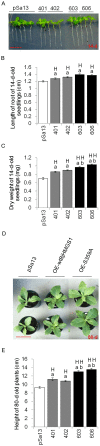
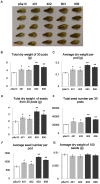
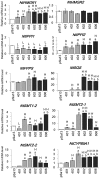
Seeds are very important not only in the life cycle of the plant but they represent food sources for man and animals. We report herein a mutant of 3-hydroxy-3-methylglutaryl-coenzyme A synthase (HMGS), the second enzyme in the mevalonate (MVA) pathway that can improve seed yield when overexpressed in a phylogenetically distant species. In Brassica juncea, the characterisation of four isogenes encoding HMGS has been previously reported. Enzyme kinetics on recombinant wild-type (wt) and mutant BjHMGS1 had revealed that S359A displayed a 10-fold higher enzyme activity. The overexpression of wt and mutant (S359A) BjHMGS1 in Arabidopsis had up-regulated several genes in sterol biosynthesis, increasing sterol content. To quickly assess the effects of BjHMGS1 overexpression in a phylogenetically more distant species beyond the Brassicaceae, wt and mutant (S359A) BjHMGS1 were expressed in tobacco (Nicotiana tabacum L. cv. Xanthi) of the family Solanaceae. New observations on tobacco OEs not previously reported for Arabidopsis OEs included: (i) phenotypic changes in enhanced plant growth, pod size and seed yield (more significant in OE-S359A than OE-wtBjHMGS1) in comparison to vector-transformed tobacco, (ii) higher NtSQS expression and sterol content in OE-S359A than OE-wtBjHMGS1 corresponding to greater increase in growth and seed yield, and (iii) induction of NtIPPI2 and NtGGPPS2 and downregulation of NtIPPI1, NtGGPPS1, NtGGPPS3 and NtGGPPS4. Resembling Arabidopsis HMGS-OEs, tobacco HMGS-OEs displayed an enhanced expression of NtHMGR1, NtSMT1-2, NtSMT2-1, NtSMT2-2 and NtCYP85A1. Overall, increased growth, pod size and seed yield in tobacco HMGS-OEs were attributed to the up-regulation of native NtHMGR1, NtIPPI2, NtSQS, NtSMT1-2, NtSMT2-1, NtSMT2-2 and NtCYP85A1. Hence, S359A has potential in agriculture not only in improving phytosterol content but also seed yield, which may be desirable in food crops. This work further demonstrates HMGS function in plant reproduction that is reminiscent to reduced fertility of hmgs RNAi lines in let-7 mutants of Caenorhabditis elegans.
Conflict of interest statement
Figures




Similar articles
-
Improved fruit α-tocopherol, carotenoid, squalene and phytosterol contents through manipulation of Brassica juncea 3-HYDROXY-3-METHYLGLUTARYL-COA SYNTHASE1 in transgenic tomato.Plant Biotechnol J. 2018 Mar;16(3):784-796. doi: 10.1111/pbi.12828. Epub 2017 Oct 17. Plant Biotechnol J. 2018. PMID: 28881416 Free PMC article.
-
Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance.Plant Biotechnol J. 2012 Jan;10(1):31-42. doi: 10.1111/j.1467-7652.2011.00631.x. Epub 2011 Jun 5. Plant Biotechnol J. 2012. PMID: 21645203
-
Overexpression and Inhibition of 3-Hydroxy-3-Methylglutaryl-CoA Synthase Affect Central Metabolic Pathways in Tobacco.Plant Cell Physiol. 2021 Mar 25;62(1):205-218. doi: 10.1093/pcp/pcaa154. Plant Cell Physiol. 2021. PMID: 33340324
-
Brassica juncea 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase 1: expression and characterization of recombinant wild-type and mutant enzymes.Biochem J. 2004 Nov 1;383(Pt. 3):517-27. doi: 10.1042/BJ20040721. Biochem J. 2004. PMID: 15233626 Free PMC article.
-
Past achievements, current status and future perspectives of studies on 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS) in the mevalonate (MVA) pathway.Plant Cell Rep. 2014 Jul;33(7):1005-22. doi: 10.1007/s00299-014-1592-9. Epub 2014 Mar 30. Plant Cell Rep. 2014. PMID: 24682521 Review.
Cited by
-
Genetic and signalling pathways of dry fruit size: targets for genome editing-based crop improvement.Plant Biotechnol J. 2020 May;18(5):1124-1140. doi: 10.1111/pbi.13318. Epub 2020 Jan 25. Plant Biotechnol J. 2020. PMID: 31850661 Free PMC article. Review.
-
Genome-wide analysis of cold imbibition stress in soybean, Glycine max.Front Plant Sci. 2023 Aug 21;14:1221644. doi: 10.3389/fpls.2023.1221644. eCollection 2023. Front Plant Sci. 2023. PMID: 37670866 Free PMC article.
-
3-Hydroxy-3-methylglutaryl coenzyme A reductase genes from Glycine max regulate plant growth and isoprenoid biosynthesis.Sci Rep. 2023 Mar 8;13(1):3902. doi: 10.1038/s41598-023-30797-4. Sci Rep. 2023. PMID: 36890158 Free PMC article.
-
Cloning and Characterization of the Gene Encoding HMGS Synthase in Polygonatum sibiricum.Biomed Res Int. 2022 Oct 7;2022:7441296. doi: 10.1155/2022/7441296. eCollection 2022. Biomed Res Int. 2022. PMID: 36246988 Free PMC article.
-
Overexpression of HMG-CoA synthase promotes Arabidopsis root growth and adversely affects glucosinolate biosynthesis.J Exp Bot. 2020 Jan 1;71(1):272-289. doi: 10.1093/jxb/erz420. J Exp Bot. 2020. PMID: 31557302 Free PMC article.
References
-
- Bach TJ (1995) Some new aspects of isoprenoid biosynthesis in plants - a review. Lipids 30: 191–202. - PubMed
-
- Hemmerlin A, Harwood JL, Bach TJ (2012) A raison d'etre for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res 51: 95–148. - PubMed
-
- Wang H, Nagegowda DA, Rawat R, Bouvier-Navé P, Guo D, et al. (2012) Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol J 10: 31–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

