Modulated expression of genes encoding estrogen metabolizing enzymes by G1-phase cyclin-dependent kinases 6 and 4 in human breast cancer cells
- PMID: 24848372
- PMCID: PMC4029737
- DOI: 10.1371/journal.pone.0097448
Modulated expression of genes encoding estrogen metabolizing enzymes by G1-phase cyclin-dependent kinases 6 and 4 in human breast cancer cells
Abstract
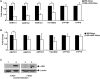
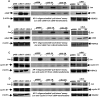
G1-phase cell cycle defects, such as alterations in cyclin D1 or cyclin-dependent kinase (cdk) levels, are seen in most tumors. For example, increased cyclin D1 and decreased cdk6 levels are seen in many human breast tumors. Overexpression of cdk6 in breast tumor cells in culture has been shown to suppress proliferation, unlike the growth stimulating effects of its close homolog, cdk4. In addition to directly affecting proliferation, alterations in cdk6 or cdk4 levels in breast tumor cells also differentially influence levels of numerous steroid metabolic enzymes (SMEs), including those involved in estrogen metabolism. Overexpression of cdk6 in tumor cell lines having low cdk6 resulted in decreased levels of mRNAs encoding aldo-keto reductase (AKR)1C1, AKR1C2 and AKR1C3, which are hydroxysteroid dehydrogenases (HSDs) involved in steroid hormone metabolism. In contrast, increasing cdk4 dramatically increased these transcript levels, especially those encoding AKR1C3, an enzyme that converts estrone to 17β-estradiol, a change that could result in a pro-estrogenic state favoring tumor growth. Effects on other estrogen metabolizing enzymes, including cytochrome P450 (CYP) 19 aromatase, 17β-HSD2, and CYP1B1 transcripts, were also observed. Interactions of cdk6 and cdk4, but not cyclin D1, with the promoter region of a cdk-regulated gene, 17β-HSD2, were detected. The results uncover a previously unsuspected link between the cell cycle and hormone metabolism and differential roles for cdk6 and cdk4 in a novel mechanism for pre-receptor control of steroid hormone action, with important implications for the origin and treatment of steroid hormone-dependent cancers.
Conflict of interest statement
Figures




Similar articles
-
Aldo-keto reductases AKR1C1, AKR1C2 and AKR1C3 may enhance progesterone metabolism in ovarian endometriosis.Chem Biol Interact. 2011 May 30;191(1-3):217-26. doi: 10.1016/j.cbi.2011.01.003. Epub 2011 Jan 11. Chem Biol Interact. 2011. PMID: 21232532
-
Aldo-keto reductase 1C3 expression in MCF-7 cells reveals roles in steroid hormone and prostaglandin metabolism that may explain its over-expression in breast cancer.J Steroid Biochem Mol Biol. 2010 Feb 15;118(3):177-87. doi: 10.1016/j.jsbmb.2009.12.009. Epub 2009 Dec 28. J Steroid Biochem Mol Biol. 2010. PMID: 20036328 Free PMC article.
-
Estrogen and progesterone metabolism in the cervix during pregnancy and parturition.J Clin Endocrinol Metab. 2008 Jun;93(6):2366-74. doi: 10.1210/jc.2007-2813. Epub 2008 Mar 25. J Clin Endocrinol Metab. 2008. PMID: 18364378 Free PMC article.
-
Structural basis of the multispecificity demonstrated by 17beta-hydroxysteroid dehydrogenase types 1 and 5.Mol Cell Endocrinol. 2006 Mar 27;248(1-2):38-46. doi: 10.1016/j.mce.2005.11.035. Epub 2006 Feb 15. Mol Cell Endocrinol. 2006. PMID: 16480815 Review.
-
Inhibitors of type 5 17β-hydroxysteroid dehydrogenase (AKR1C3): overview and structural insights.J Steroid Biochem Mol Biol. 2011 May;125(1-2):95-104. doi: 10.1016/j.jsbmb.2010.11.004. Epub 2010 Nov 16. J Steroid Biochem Mol Biol. 2011. PMID: 21087665 Free PMC article. Review.
Cited by
-
siRNA Delivery for Control of Cyclin D1 and E2F1 Expression in Crohn's Disease.Transl Med UniSa. 2018 Mar 31;17:25-33. eCollection 2017 Jul. Transl Med UniSa. 2018. PMID: 30083520 Free PMC article.
-
siRNA Delivery for Control of Cyclin D1 and E2F1 Expression in Crohn's Disease.Transl Med UniSa. 2018 Mar 31;17:22-30. eCollection 2017 Jul. Transl Med UniSa. 2018. PMID: 30050877 Free PMC article.
-
New targeted therapies for breast cancer: A focus on tumor microenvironmental signals and chemoresistant breast cancers.World J Clin Cases. 2014 Dec 16;2(12):769-86. doi: 10.12998/wjcc.v2.i12.769. World J Clin Cases. 2014. PMID: 25516852 Free PMC article. Review.
-
Non-canonical roles of PFKFB3 in regulation of cell cycle through binding to CDK4.Oncogene. 2018 Mar;37(13):1685-1698. doi: 10.1038/s41388-017-0072-4. Epub 2018 Jan 16. Oncogene. 2018. PMID: 29335521
-
Cyclin-Dependent Kinases 4/6 Inhibitors in Breast Cancer: Current Status, Resistance, and Combination Strategies.J Cancer. 2019 Aug 29;10(22):5504-5517. doi: 10.7150/jca.32628. eCollection 2019. J Cancer. 2019. PMID: 31632494 Free PMC article.
References
-
- Ho A, Dowdy SF (2002) Regulation of (G)1 cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genet Dev 12: 47–52. - PubMed
-
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG (2004) Cyclin D1: normal and abnormal functions. Endocrinol 145: 5439–5447. - PubMed
-
- Yu Q, Sicinska E, Geng Y, Ahnstrom M, Zagozdzon A, et al. (2006) Requirement for cdk4 kinase function in breast cancer. Cancer Cell 9: 23–32. - PubMed
-
- Grossel MJ, Hinds PW (2006) Beyond the cell cycle: A new role for cdk6 in differentiation. J Cell Biochem 97: 485–493. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

