Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation
- PMID: 24848695
- PMCID: PMC4107114
- DOI: 10.1002/humu.22594
Genetic variations and diseases in UniProtKB/Swiss-Prot: the ins and outs of expert manual curation
Abstract
During the last few years, next-generation sequencing (NGS) technologies have accelerated the detection of genetic variants resulting in the rapid discovery of new disease-associated genes. However, the wealth of variation data made available by NGS alone is not sufficient to understand the mechanisms underlying disease pathogenesis and manifestation. Multidisciplinary approaches combining sequence and clinical data with prior biological knowledge are needed to unravel the role of genetic variants in human health and disease. In this context, it is crucial that these data are linked, organized, and made readily available through reliable online resources. The Swiss-Prot section of the Universal Protein Knowledgebase (UniProtKB/Swiss-Prot) provides the scientific community with a collection of information on protein functions, interactions, biological pathways, as well as human genetic diseases and variants, all manually reviewed by experts. In this article, we present an overview of the information content of UniProtKB/Swiss-Prot to show how this knowledgebase can support researchers in the elucidation of the mechanisms leading from a molecular defect to a disease phenotype.
Keywords: UniProtKB/Swiss-Prot; controlled vocabulary; database; disease; functional annotation; genetic variants; manual curation.
© 2014 The Authors. *Human Mutation published by Wiley Periodicals, Inc.
Figures
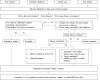
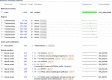
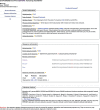

References
-
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, Lupski JR, Katsanis N. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72:1187–1199. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5R01GM080646-07/GM/NIGMS NIH HHS/United States
- R01 GM080646/GM/NIGMS NIH HHS/United States
- 8P20GM103446-12/GM/NIGMS NIH HHS/United States
- 5G08LM010720-03/LM/NLM NIH HHS/United States
- 4U41 HG006104-04/HG/NHGRI NIH HHS/United States
- 2P41 HG02273/HG/NHGRI NIH HHS/United States
- P20 GM103446/GM/NIGMS NIH HHS/United States
- U41 HG006104/HG/NHGRI NIH HHS/United States
- U41 HG007822/HG/NHGRI NIH HHS/United States
- P41 HG002273/HG/NHGRI NIH HHS/United States
- G08 LM010720/LM/NLM NIH HHS/United States
- 3R01GM080646-07S1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

