Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes
- PMID: 24850747
- PMCID: PMC4136290
- DOI: 10.1128/JVI.00531-14
Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes
Abstract
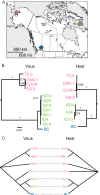
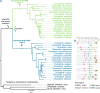
Bunyaviridae is a large family of RNA viruses chiefly comprised of vertebrate and plant pathogens. We discovered novel bunyavirids that are approximately equally divergent from each of the five known genera. We characterized novel genome sequences for two bunyavirids, namely, Kigluaik phantom virus (KIGV), from tundra-native phantom midges (Chaoborus), and Nome phantom virus (NOMV), from tundra-invading phantom midges, and demonstrated that these bunyavirid-like sequences belong to an infectious virus by passaging KIGV in mosquito cell culture, although the infection does not seem to be well sustained beyond a few passages. Virus and host gene sequences from individuals collected on opposite ends of North America, a region spanning 4,000 km, support a long-term, vertically transmitted infection of KIGV in Chaoborus trivittatus. KIGV-like sequences ranging from single genes to full genomes are present in transcriptomes and genomes of insects belonging to six taxonomic orders, suggesting an ancient association of this clade with insect hosts. In Drosophila, endogenous virus genes have been coopted, forming an orthologous tandem gene family that has been maintained by selection during the radiation of the host genus. Our findings indicate that bunyavirid-host interactions in nonbloodsucking arthropods have been much more extensive than previously thought.
Importance: Very little is known about the viral diversity in polar freshwater ponds, and perhaps less is known about the effects that climate-induced habitat changes in these regions will have on virus-host interactions in the coming years. Our results show that at the tundra-boreal boundary, a hidden viral landscape is being altered as infected boreal phantom midges colonize tundra ponds. Likewise, relatively little is known of the deeper evolutionary history of bunyavirids that has led to the stark lifestyle contrasts between some genera. The discovery of this novel bunyavirid group suggests that ancient and highly divergent bunyavirid lineages remain undetected in nature and may offer fresh insight into host reservoirs, potential sources of emerging disease, and major lifestyle shifts in the evolutionary history of viruses in the family Bunyaviridae.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Jorgenson MT, Shur YL, Pullman ER. 2006. Abrupt increase in permafrost degradation in Arctic Alaska. Geophys. Res. Lett. 33:L02503. 10.1029/2005GL024960 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

