Evolution of the Phosphoenolpyruvate Carboxylase Protein Kinase Family in C3 and C4 Flaveria spp
- PMID: 24850859
- PMCID: PMC4081323
- DOI: 10.1104/pp.114.240283
Evolution of the Phosphoenolpyruvate Carboxylase Protein Kinase Family in C3 and C4 Flaveria spp
Abstract
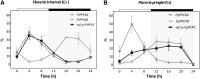
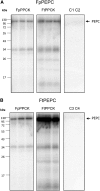
The key enzyme for C4 photosynthesis, Phosphoenolpyruvate Carboxylase (PEPC), evolved from nonphotosynthetic PEPC found in C3 ancestors. In all plants, PEPC is phosphorylated by Phosphoenolpyruvate Carboxylase Protein Kinase (PPCK). However, differences in the phosphorylation pattern exist among plants with these photosynthetic types, and it is still not clear if they are due to interspecies differences or depend on photosynthetic type. The genus Flaveria contains closely related C3, C3-C4 intermediate, and C4 species, which are evolutionarily young and thus well suited for comparative analysis. To characterize the evolutionary differences in PPCK between plants with C3 and C4 photosynthesis, transcriptome libraries from nine Flaveria spp. were used, and a two-member PPCK family (PPCKA and PPCKB) was identified. Sequence analysis identified a number of C3- and C4-specific residues with various occurrences in the intermediates. Quantitative analysis of transcriptome data revealed that PPCKA and PPCKB exhibit inverse diel expression patterns and that C3 and C4 Flaveria spp. differ in the expression levels of these genes. PPCKA has maximal expression levels during the day, whereas PPCKB has maximal expression during the night. Phosphorylation patterns of PEPC varied among C3 and C4 Flaveria spp. too, with PEPC from the C4 species being predominantly phosphorylated throughout the day, while in the C3 species the phosphorylation level was maintained during the entire 24 h. Since C4 Flaveria spp. evolved from C3 ancestors, this work links the evolutionary changes in sequence, PPCK expression, and phosphorylation pattern to an evolutionary phase shift of kinase activity from a C3 to a C4 mode.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures




Similar articles
-
Evolution of c4 phosphoenolpyruvate carboxylase. Genes and proteins: a case study with the genus Flaveria.Ann Bot. 2004 Jan;93(1):13-23. doi: 10.1093/aob/mch003. Epub 2003 Nov 26. Ann Bot. 2004. PMID: 14644912 Free PMC article. Review.
-
Evolution of C4 phosphoenolpyruvate carboxylase.Arch Biochem Biophys. 2003 Jun 15;414(2):180-8. doi: 10.1016/s0003-9861(03)00165-6. Arch Biochem Biophys. 2003. PMID: 12781769 Review.
-
Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria--a gradual increase from C3 to C4 characteristics.Planta. 2003 Sep;217(5):717-25. doi: 10.1007/s00425-003-1045-0. Epub 2003 Jun 13. Planta. 2003. PMID: 12811556
-
Dynamic changes of genome sizes and gradual gain of cell-specific distribution of C4 enzymes during C4 evolution in genus Flaveria.Plant Genome. 2021 Jul;14(2):e20095. doi: 10.1002/tpg2.20095. Epub 2021 Apr 29. Plant Genome. 2021. PMID: 33913619
-
Mesophyll Chloroplast Investment in C3, C4 and C2 Species of the Genus Flaveria.Plant Cell Physiol. 2016 May;57(5):904-18. doi: 10.1093/pcp/pcw015. Epub 2016 Mar 16. Plant Cell Physiol. 2016. PMID: 26985020
Cited by
-
The limiting factors and regulatory processes that control the environmental responses of C3, C3-C4 intermediate, and C4 photosynthesis.Oecologia. 2021 Dec;197(4):841-866. doi: 10.1007/s00442-021-05062-y. Epub 2021 Oct 29. Oecologia. 2021. PMID: 34714387 Free PMC article. Review.
-
Identification and evolution of C4 photosynthetic pathway genes in plants.BMC Plant Biol. 2020 Mar 30;20(1):132. doi: 10.1186/s12870-020-02339-x. BMC Plant Biol. 2020. PMID: 32228460 Free PMC article.
-
Molecular Cloning of Novel-Type Phosphoenolpyruvate Carboxylase Isoforms in Pitaya (Hylocereus undatus).Plants (Basel). 2020 Sep 21;9(9):1241. doi: 10.3390/plants9091241. Plants (Basel). 2020. PMID: 32967083 Free PMC article.
-
Regulation of Crassulacean acid metabolism at the protein level in Kalanchoë laxiflora.Plant Physiol. 2025 Mar 28;197(4):kiaf095. doi: 10.1093/plphys/kiaf095. Plant Physiol. 2025. PMID: 40259462 Free PMC article.
-
Transcriptional Profiling and Identification of Heat-Responsive Genes in Perennial Ryegrass by RNA-Sequencing.Front Plant Sci. 2017 Jun 21;8:1032. doi: 10.3389/fpls.2017.01032. eCollection 2017. Front Plant Sci. 2017. PMID: 28680431 Free PMC article.
References
-
- Agetsuma M, Furumoto T, Yanagisawa S, Izui K. (2005) The ubiquitin-proteasome pathway is involved in rapid degradation of phosphoenolpyruvate carboxylase kinase for C4 photosynthesis. Plant Cell Physiol 46: 389–398 - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 - PubMed
-
- Aubry S, Brown NJ, Hibberd JM. (2011) The role of proteins in C(3) plants prior to their recruitment into the C(4) pathway. J Exp Bot 62: 3049–3059 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

