Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers
- PMID: 24850867
- PMCID: PMC4060725
- DOI: 10.1073/pnas.1407688111
Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers
Abstract
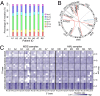
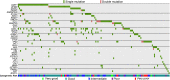
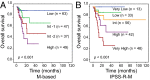
Myelodysplastic syndrome (MDS) includes a group of diseases characterized by dysplasia of bone marrow myeloid lineages with ineffective hematopoiesis and frequent evolution to acute myeloid leukemia (AML). Whole-genome sequencing was performed in CD34(+) hematopoietic stem/progenitor cells (HSPCs) from eight cases of refractory anemia with excess blasts (RAEB), the high-risk subtype of MDS. The nucleotide substitution patterns were found similar to those reported in AML, and mutations of 96 protein-coding genes were identified. Clonal architecture analysis revealed the presence of subclones in six of eight cases, whereas mutation detection of CD34(+) versus CD34(-) cells revealed heterogeneity of HSPC expansion status. With 39 marker genes belonging to eight functional categories, mutations were analyzed in 196 MDS cases including mostly RAEB (n = 89) and refractory cytopenia with multilineage dysplasia (RCMD) (n = 95). At least one gene mutation was detected in 91.0% of RAEB, contrary to that in RCMD (55.8%), suggesting a higher mutational burden in the former group. Gene abnormality patterns differed between MDS and AML, with mutations of activated signaling molecules and NPM1 being rare, whereas those of spliceosome more common, in MDS. Finally, gene mutation profiles also bore prognostic value in terms of overall survival and progression free survival.
Keywords: clonal evolution; gene mutation pattern; prognostic stratification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361(19):1872–1885. - PubMed
-
- Greenberg PL, Young NS, Gattermann N. Myelodysplastic syndromes. Hematology (Am Soc Hematol Educ Program) 2002:136–161. - PubMed
-
- Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114(5):937–951. - PubMed
-
- Greenberg P, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89(6):2079–2088. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP0141438
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

