Structure and function of atypically coordinated enzymatic mononuclear non-heme-Fe(II) centers
- PMID: 24850951
- PMCID: PMC4019311
- DOI: 10.1016/j.ccr.2012.04.028
Structure and function of atypically coordinated enzymatic mononuclear non-heme-Fe(II) centers
Abstract
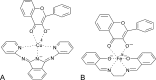
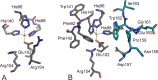
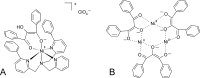
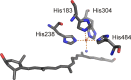
Mononuclear, non-heme-Fe(II) centers are key structures in O2 metabolism and catalyze an impressive variety of enzymatic reactions. While most are bound via two histidines and a carboxylate, some show a different organization. A short overview of atypically coordinated O2 dependent mononuclear-non-heme-Fe(II) centers is presented here Enzymes with 2-His, 3-His, 3-His-carboxylate and 4-His bound Fe(II) centers are discussed with a focus on their reactivity, metal ion promiscuity and recent progress in the elucidation of their enzymatic mechanisms. Observations concerning these and classically coordinated Fe(II) centers are used to understand the impact of the metal binding motif on catalysis.
Keywords: 1,3-bis(2-pyridylimino)isoindoline, ind; 2OH-1,3-Ph2PD, 2-hydroxy-1,3-diphenylpropanedione; 6-Ph2TPA, N,N-bis[(6-phenyl-2-pyridyl)methyl]-N-[(2-pyridyl)-methyl]amine; ADO, cysteamine dioxygenase; AO, apocarotenoid 15,15′-oxygenase; ARD, aci-reductone dioxygenase; BsQDO, quercetin 2,3-dioxygenase from Bacillus subtilis; CD, circular dichroism; CDO, cysteine dioxygenase; CGDO, 5-chloro-gentisate 1,2-dioxygenase; CS2, clavaminate synthase; CarOs, carotenoid oxygenases; DFT, density functional theory; Dioxygen activation; Dioxygenase; Dke1, diketone dioxygenase; EPR, electron paramagnetic resonance; EXAFS, extended X-ray absorption fine structure spectroscopy; Enzyme catalysis; Facial triad; GDO, gentisate 1,2-dioxygenase; HADO, 3-hydroxyanthranilate 3,4-dioxygenase; HGDO, homogentisate 1,2-dioxygenase; HNDO, hydroxy-2-naphthoate dioxygenase; MCD, magnetic circular dichroism; MNHEs, mononuclear non-heme-Fe(II) dependent enzymes; Metal binding motif; NRP, nonribosomal peptide; OTf-, trifluormethanesulfonate; PDB, protein data bank; QDO, quercetin 2,3-dioxygenase; SDO, salicylate 1,2-dioxygenase; Structure–function relationships; TauD, taurine hydroxylase; XAS, X-ray absorption spectroscopy; acac, acetylacetone (2,4-pentanedione); fla, flavonolate; α-KG, α-ketoglutarate.
Figures
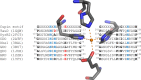

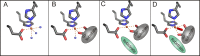
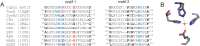
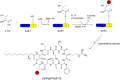
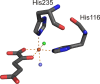

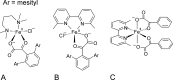



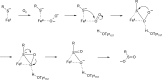
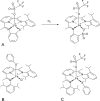
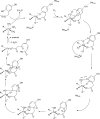
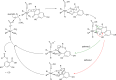
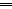 O is observed instead.
O is observed instead.
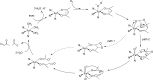

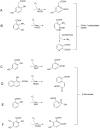

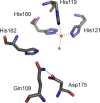




References
-
- Solomon E.I., Brunold T.C., Davis M.I., Kemsley J.N., Lee S.K., Lehnert N., Neese F., Skulan A.J., Yang Y.S., Zhou J. Chem. Rev. 2000;100:235. - PubMed
-
- Costas M., Mehn M.P., Jensen M.P., Que L., Jr. Chem. Rev. 2004;104:939. - PubMed
-
- Schenk G., Pau M.Y., Solomon E.I. J. Am. Chem. Soc. 2004;126:505. - PubMed
-
- Neidig M.L., Solomon E.I. Chem. Commun. 2005:5843. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
